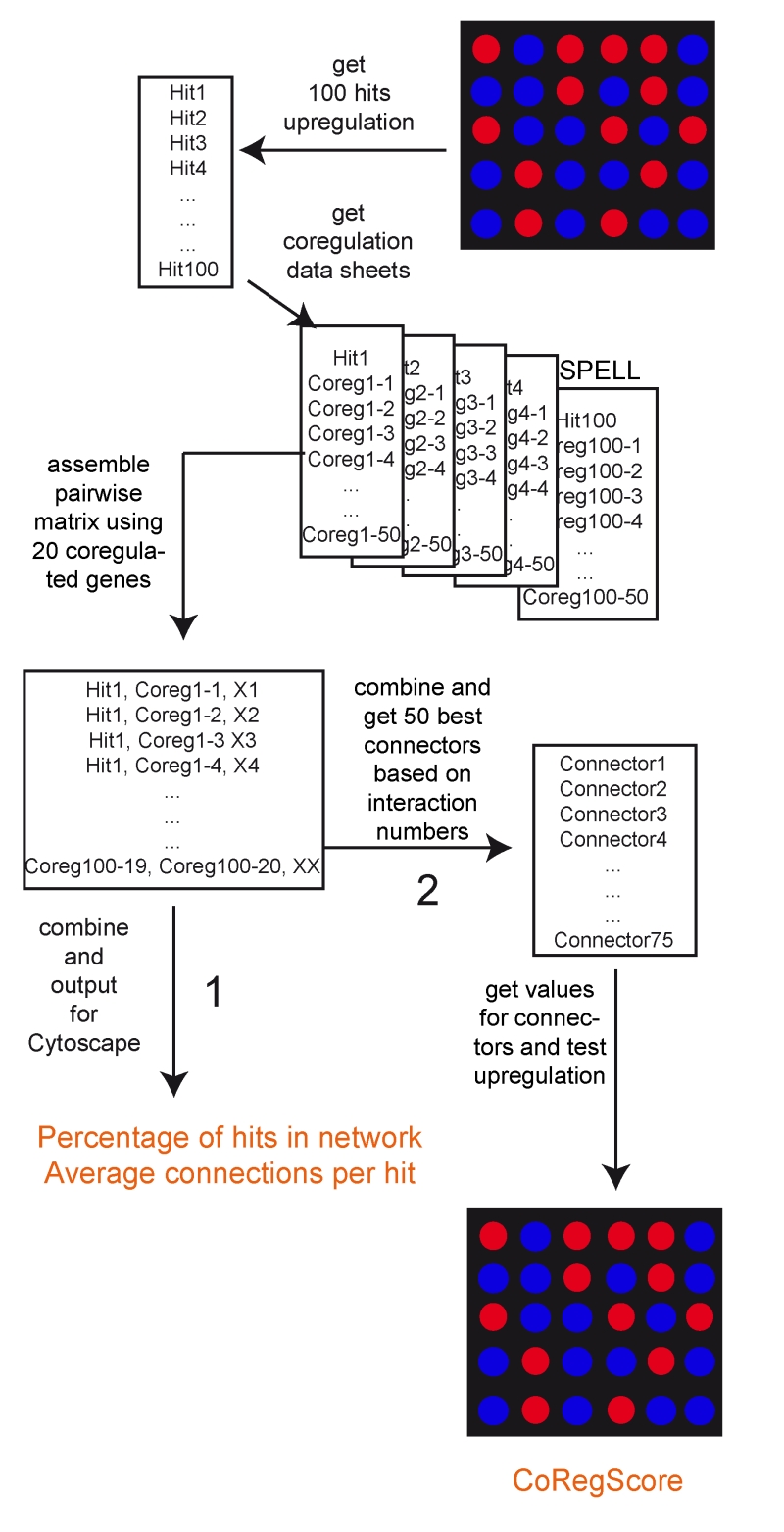
FIGURE 1: Data processing in ClusterEx. A flexible number of hits (here the top 100) are obtained from the microarray data set as “hits”. For each hit, the best co-regulators (here 50) are obtained from the SPELL database. These are assembled into a pairwise matrix. After combining, each pair is only listed once with the number of occurrences (X). This matrix then is exported to Cytoscape and values are reported for percentage of hits included in the network and connection numbers per hit (Pathway 1). Furthermore, a flexible number (here 50) of connectors from the matrix can be included into the network. For those the real expression values are obtained from the experimental data set. The CoRegScore is calculated from the positioning of these predicted connectors in the hit list of the experiment (Pathway 2). This procedure is described in the materials and methods section.
By continuing to use the site, you agree to the use of cookies. more information
The cookie settings on this website are set to "allow cookies" to give you the best browsing experience possible. If you continue to use this website without changing your cookie settings or you click "Accept" below then you are consenting to this. Please refer to our "privacy statement" and our "terms of use" for further information.