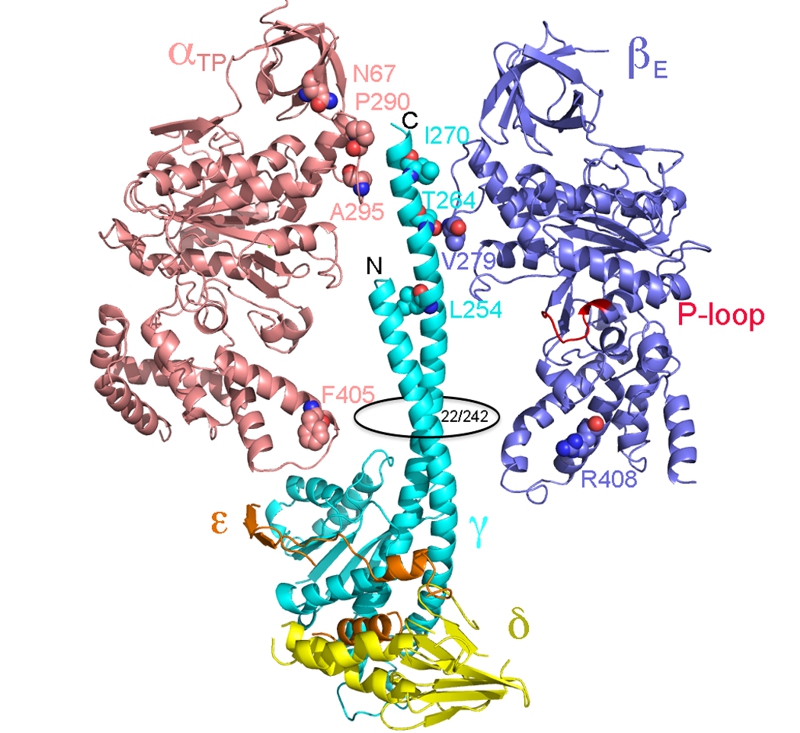
FIGURE 3: Location of residues in the yeast ATP synthase classified as mgi.
The structural representation is derived from the x-ray structure of the yeast F1 ATPase (2HDL). Shown are the αβγδε subunits, as indicated. The α3β3 core has been stripped down to a single α and β subunit to simplify the view. The mgi residues are as labeled. The N- and C- terminal ends of the γ-subunit are indicated as is the region of the orifice of the α3β3 assembly, which is located at residues 22 and 242 of the yeast γ-subunit. The nucleotide binding P-loop domain is colored red and labeled.