Back to article: Urm1, not quite a ubiquitin-like modifier?
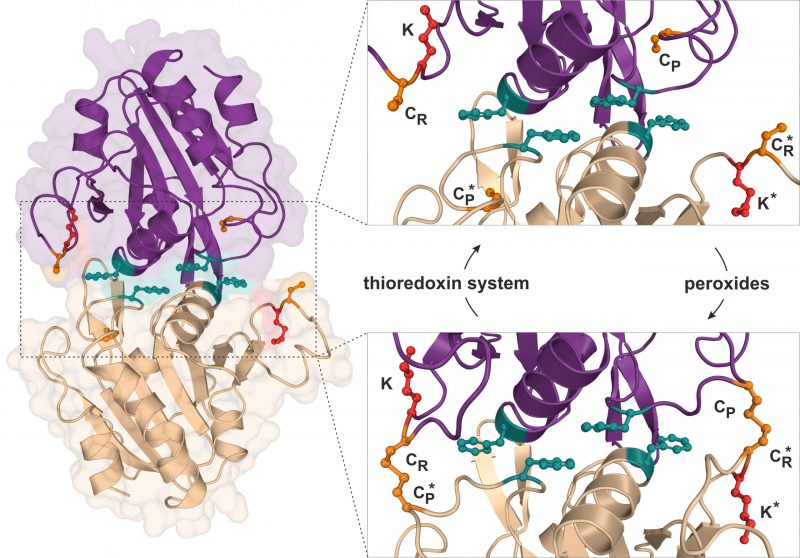
FIGURE 2: Structure and catalytic cycle of the peroxiredoxin Ahp1 from S. cerevisiae. Ribbon diagram presentation of the dimeric Ahp1 enzyme with its two subunits (magenta & wheat) in its reduced form (PDB: 4DSR). Residues critical for peroxidase activity (orange), dimerization (teal) and Urm1 conjugation (red) are highlighted. The enlarged insert focusses on the hydrophobic homodimer interface. In its reduced form (top panel), the peroxidatic cysteine (Cys-62: CP) is buried inside the active center. Upon oxidation by peroxides, the CP approaches the resolving cysteine of the opposite subunit (Cys-31: CR*) leading to formation of intersubunit disulfide bridges (bottom panel, PDB: 4DSQ). These are subsequently reduced by the thioredoxin system. Note that next to each CR is a lysine residue (Lys-32: K) previously reported to function as the sole site for Ahp1 urmylation [19].
19. Van der Veen AG, Schorpp K, Schlieker C, Buti L, Damon JR, Spooner E, Ploegh H, and Jentsch S (2011). Role of the ubiquitin-like protein Urm1 as a noncanonical lysine-directed protein modifier. Proc Natl Acad Sci USA 108(5): 1763–1770. 10.1073/pnas.1014402108