Back to article: Exploring absent protein function in yeast: assaying post translational modification and human genetic variation
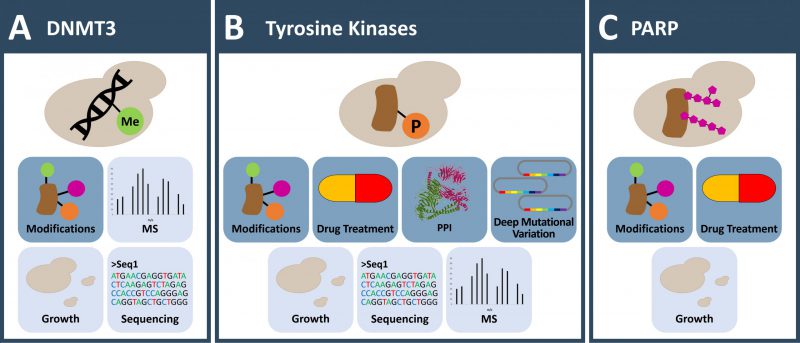
FIGURE 1: Applications to investigate DNA methylation and post translational protein modifications otherwise absent in yeast. (A) Yeast is used as a model organism to investigate cytosine DNA methylation induced de novo via human DNMTs. A special focus lies on the interplay with histone modifications. (B) Tyrosine kinases are lacking in yeasts and their activity can be toxic. This allows the investigation of protein variants (in deep mutagenesis screens) that govern kinase activity and function and enables inhibitor testing. Non-toxic low-level expression of human tyrosine kinases in yeast allows large scale screening of phosphorylation dependent protein interaction and the characterization of kinase substrate specificity. (C) PARPs attach ADP-ribose to their target proteins. No such modification is reported in yeast. Expression of PARPs in yeast results in a growth phenotype and allows screening for substrates and small molecule inhibitory drugs. (A-C) Icons with dark blue background: observable; Icons with light blue background: readout; MS: mass spectrometry; PPI: protein-protein-interaction.