Reviews:
Microbial Cell, Vol. 5, No. 11, pp. 482 - 494; doi: 10.15698/mic2018.11.655
Conventional and emerging roles of the energy sensor Snf1/AMPK in Saccharomyces cerevisiae
1 Department of Biotechnology and Biosciences, University of Milano-Bicocca, Milan, Italy.
2 SYSBIO, Centre of Systems Biology, Milan, Italy.
3 Present address: Department of Biology, University of Fribourg, Fribourg, Switzerland.
Keywords: budding yeast, metabolism, stress response, aging, transcription, signaling, cell cycle, endocytosis, DNA damage, glucose repression.
Abbreviations:
AIS - autoinhibitory sequence.
Received originally: 29/06/2018 Received in revised form: 23/08/2018
Accepted: 23/08/2018
Published: 29/09/2018
Correspondence:
Paola Coccetti, Department of Biotechnology and Biosciences, University of Milano-Bicocca, Milan, Italy; paola.coccetti@unimib.it
Conflict of interest statement: The authors have no conflicts of interest to declare.
Please cite this article as: Paola Coccetti, Raffaele Nicastro and Farida Tripodi (2018). Conventional and emerging roles of the energy sensor Snf1/AMPK in Saccharomyces cerevisiae. Microbial Cell: in press.
Abstract
All proliferating cells need to match metabolism, growth and cell cycle progression with nutrient availability to guarantee cell viability in spite of a changing environment. In yeast, a signaling pathway centered on the effector kinase Snf1 is required to adapt to nutrient limitation and to utilize alternative carbon sources, such as sucrose and ethanol. Snf1 shares evolutionary conserved functions with the AMP-activated Kinase (AMPK) in higher eukaryotes which, activated by energy depletion, stimulates catabolic processes and, at the same time, inhibits anabolism. Although the yeast Snf1 is best known for its role in responding to a number of stress factors, in addition to glucose limitation, new unconventional roles of Snf1 have recently emerged, even in glucose repressing and unstressed conditions. Here, we review and integrate available data on conventional and non-conventional functions of Snf1 to better understand the complexity of cellular physiology which controls energy homeostasis.
INTRODUCTION
Cell growth and proliferation require a high amount of energy for biosynthetic pathways. Cells take energy from nutrient intake and both unicellular and multicellular eukaryotes have evolved systems that allow dynamic sensing of energy sources, mainly sugars. The class of Snf1/AMPK (Sucrose non-fermenting/AMP-activated protein kinase) plays a key role as a guardian of cellular energy [1]. They are highly conserved serine/threonine kinases and their primary role is the integration of signals regarding nutrient availability and environmental stress, ensuring the adaptation to those conditions and cell survival [2].
–
Here we discuss the mechanisms of action of Snf1, a member of the Snf1/AMPK family in Saccharomyces cerevisiae and its conventional roles in the regulation of metabolism, stress response and aging. In addition, we also focus on recent advances showing emerging functions of Snf1 on the modulation of key processes such as endocytosis and cellular trafficking as well as cell cycle, proliferation and metabolism.
SNF1 COMPLEX COMPOSITION
Protein kinase Snf1 in yeast is a heterotrimeric complex made by the catalytic α subunit Snf1, a regulatory β subunit (alternatively Gal83, Sip1 and Sip2) and the γ subunit Snf4 [3].
The catalytic α subunit (encoded by the SNF1 gene) was identified in a screening of mutants unable to grow in presence of sucrose as carbon source [4]. The Snf1 subunit is constitutively expressed and constituted by a catalytic N-terminal domain and a C-terminal regulatory region. The regulatory region presents a short autoinhibitory sequence (AIS) (380 – 415 aa) and a domain which mediates the interactions with the β subunits of the complex. The autoinhibitory domain interacts with both the regulatory subunit Snf4 and the kinase domain of Snf1. The interaction with Snf4 relieves the inhibition of the AIS allowing the phosphorylation of Thr210 residue of Snf1 that determines its activation [5][6].
–
In S. cerevisiae, three β subunits (Gal83, Sip1 and Sip2) are present. They share partially redundant functions, since only the triple mutant sip1Δsip2Δgal83Δ strain shows growth defects when glycerol or ethanol are added as carbon sources [7][8]. The β subunits contain a conserved C-terminal sequence in which two domains are present: the KIS domain (Kinase Interacting Sequence) that mediates the interaction with the α-subunit Snf1 [9] and the ASC domain (Association with SNF1 kinase Complex) that allows the interaction with Snf4 [10]. Differently, the N-terminal sequence is specific for each β subunit and confers a different subcellular localization pattern to each protein. All three proteins are mainly cytoplasmic in presence of high glucose concentrations. Upon glucose depletion, Sip1 relocalizes to the vacuolar membrane, Gal83 relocalizes to the nucleus, while Sip2 remains cytoplasmic [11]. Thus, the role of the β subunits is to interact with Snf1 and to modulate its subcellular localization [11][12]. The particular localization of the kinase complexes with different β subunits confers specialized functions. For example, Sip1 alone is not able to sustain growth on ethanol or glycerol and determines a very low kinase activity of the complex [13], Sip2 function seems to be involved in the mechanism of cellular aging [14], while Gal83 plays its main role in the Snf1-dependent transcriptional regulation, since in low glucose it determines the nuclear localization of the Snf1 complex thank to its NLS (Nuclear Localization Signal). On the contrary, NES (Nuclear Export Signal), present on the sequence of Gal83, allows the exit from the nucleus of the complex when high glucose concentrations are available [15]. In addition, Gal83 mediates the interaction of Snf1 with some substrates, such as the transcription activator Sip4 [16] and the transcriptional apparatus [17]. It has also been shown that deletion of the glycogen binding domain (GBD) of Gal83 leads to a constitutive activation of Snf1, which results able to modulate the expression of some Snf1-regulated genes also in high glucose concentrations [18]. The GBD domain of Gal83 also interacts with the Reg1/Glc7 phosphatase complex, responsible for Snf1 inactivation [18]. Taken together those data suggest that Gal83 plays a dual role regulating nuclear localization of Snf1 in low glucose and guaranteeing its inactivation in high glucose.
–
Similarly to SNF1, the gene encoding the γ subunit, SNF4, was identified by isolation of a sucrose non fermenting mutant [4]. Snf4 is a constitutively expressed protein that binds both the α and β subunits of the Snf1 complex [10][19]. The role of Snf4 is to relieve the inhibition of Snf1 interacting with its AIS domain, stabilizing the Snf1 complex in the active conformation [20]. In fact, SNF4 deletion causes a decreased kinase activity of Snf1, whereas deletion of the AIS domain of Snf1 fully complement the phenotype of a snf4Δ strain [19][20]. Remarkably, the activating phosphorylation of Thr210 residue of Snf1 is still detectable in a snf4Δ strain [21] and in high glucose Snf4 seems to be required for the proper inactivation of Snf1 mediated by the phosphatase complex Reg1/Glc7 [18]. Thus, these findings indicate that Snf4 plays a complex role in the regulation of Snf1.
REGULATION OF SNF1 ACTIVITY
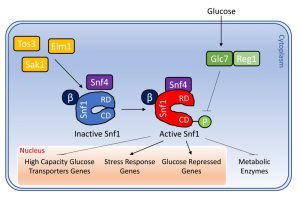
Snf1 complex is activated through phosphorylation of the Thr210 of the α subunit by one of the three constitutively active upstream kinases Sak1, Tos3 and Elm1 [22][23] (Fig. 1). This phosphorylation is essential for Snf1 activity, since the sak1Δtos3Δelm1Δ strain shows the same phenotype of a snf1Δ strain, such as growth defects in presence of limiting glucose or alternative carbon sources like glycerol or ethanol [22].
–
Although Snf1 phosphorylation is a key step for its activation, a non-phosphorylatable Snf1 mutant (Snf1-T210A) retains a low catalytic activity, originating intermediate phenotypes [24][25]. Also, the mutation of the lysine which constitutes the ATP binding site in the kinase domain (Snf1-K84R), which for many aspects mimics the loss of Snf1 protein, still confers a slight catalytic activity [26][27][28][29].
–
On the other side, in response to high glucose concentrations Snf1 is inactivated through dephosphorylation of Thr210 by the Gcl7 protein phosphatase (also known as PP1), which is targeted to Snf1 by the adaptor subunit Reg1 [30][31]. Reg1 interacts both with Glc7 and Snf1 when glucose is largely available in the culture medium and loss of Reg1 leads to the constitutive activation of Snf1 [32][33][34]. It has been reported that in high glucose concentration, Hxk2 (Hexokinase 2) regulates the activity of PP1 and consequently the activation of Snf1 kinase [31].
–
Active Snf1 phosphorylates serine and threonine residues contained in the consensus pattern Φ-x-R-x-x-S/T-x-x-x-Φ, where Φ is a hydrophobic residue [35].
–
Differently from its mammalian homolog AMPK, yeast protein kinase Snf1 is not allosterically activated by AMP [36]. However, it was demonstrated that ADP molecules are able to bind the γ subunit Snf4, preventing Snf1 dephosphorylation mediated by Glc7 [37][38].
–
The structure of the kinase domain of Snf1 showed that it is a dimer, which represents an inactive form of the kinase, since Thr210 is inaccessible for phosphorylation by the activating kinases [39]. Although these results suggest the existence of another layer of regulation of Snf1 activity, further investigation is required to better elucidate its physiological relevance.
–
Interestingly, some recent evidence indicates additional mechanisms that regulate Snf1 activity: (i) phosphorylation of Ser214, inside the activation loop, downregulates Snf1 function [40]; (ii) SUMOylation of the catalytic α subunit Snf1 inhibits its activity, possibly by attenuating its levels in the cell and/or favoring the inactive conformation of the kinase [41]; (iii) the SAGA acetyl transferase complex deubiquitylates Snf1 affecting the stability of the complex and its kinase activity [42]; (iv) the ubiquitin-associated motif (UBA) of the α subunit Snf1 indirectly regulates SNF1 gene expression and Snf1 interaction with the γ subunit Snf4 [43].
NETWORK OF SNF1 PHYSICAL INTERACTORS
A total of 216 proteins physically interacting with Snf1 are annotated in SGD (Saccharomyces Genome Database, http://www.yeastgenome.org), 92 of which are also Snf1 substrates (identified by high throughput or low throughput assays). In Figure 2 we clustered them on the base of their function. Apart from the most known interactors involved in Snf1 complex regulation, transcription, histone modification, signaling and metabolism, there are many proteins which regulate translation, ribosome function, intracellular transport/trafficking and cell cycle. In addition, some of them are also proteins of the ubiquitin/proteasome machinery, chaperones and Fe/S cluster proteins (Fig. 2). Remarkably, only a few of them have been extensively investigated for the physiological relevance of Snf1-dependent phosphorylations, suggesting that many functions of Snf1 are still to be discovered.
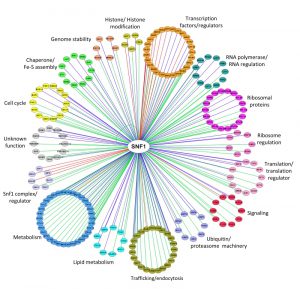 | FIGURE 2: Network of Snf1 physical interactors. The network reports the known physical associations obtained from SGD (Saccharomyces Genome Database, http://www.yeastgenome.org). Interactors are clustered according to their function and colored differently. When the interactor is also a substrate of Snf1 according to the Yeast Kinase Interaction Database (KID, http://www.moseslab.csb.utoronto.ca/KID/); [44]), the edge is colored in red if phosphorylation was analyzed by low throughput assays (LTP in vitro kinase assays; in vitro phosphorylation site mapping; in vivo phosphorylation site mapping; phosphorylation reduced or absent in kinase mutant) or in blue if phosphorylation was assayed only by high throughput analysis (protein chip data for in vitro phosphorylated substrate; HTP in vitro phosphorylation). Data visualization and analysis was performed with Cytoscape [45]. |
CONVENTIONAL ROLES OF SNF1
Snf1 and the regulation of transcription
The most studied function of Snf1 is the regulation of transcription, involving more than 400 genes [44]. Snf1 acts both on transcription factors and on chromatin remodeling [45][46][47][48], as also highlighted by the number of its interactors belonging to these two classes of proteins (Fig. 2).
–
Mig1 is the most important glucose-regulated transcriptional repressor [49]. Mig1 is phosphorylated by Snf1 on four sites when glucose is scarce, causing the activation of a NES (Nuclear Export Signal) sequence that causes its translocation from the nucleus to the cytoplasm through the exportin Msn5 [46][50][51]. Important in the regulation of Mig1 is hexokinase Hxk2, which interacts with the transcriptional repressor directly in the nucleus to avoid its phosphorylation by Snf1, thus providing a link between glucose metabolism and transcription of glucose-repressed genes [52]. Mig1 represses about 90 genes, including those coding for enzymes required for the metabolism of sucrose (SUC2), maltose (the MAL regulon) and galactose (GAL4) [53]. Furthermore, Mig1 controls the expression of high-affinity glucose transporters, required when glucose is scarce (HXT2, HXT4) [54], represses TPS1, essential for the metabolism of trehalose [55] and genes coding for enzymes of the TCA cycle [56].
–
Besides Mig1, Snf1 regulates the activity of other transcription factors. Cat8 and Sip4, which bind Carbon Source Responsive Elements (CSRE), regulate the expression of gluconeogenic genes [57] and are activated by Snf1 phosphorylation [47][58]. Cat8 activates the expression of glucose-repressed genes alongside transcription factor Adr1, which is itself a target of Snf1 [59][60]. Moreover, in a fine mechanism of positive feedback, the CAT8 gene is activated by Snf1 through inhibition of Mig1 [45]. In addition, Gcn4, the transcription factor responsible for the expression of genes involved in amino acid biosynthesis, is also regulated by Snf1 when in complex with the β subunit Gal83 or Sip1, but not Sip2 [61].
–
Snf1 has been reported to phosphorylate Ser10 of histone H3 and to promote the acetylation on Lys14 of histone H3 by Gcn5, a component of the SAGA complex [48]. Snf1-mediated regulation of histone H3 is involved in the expression of ADY2 gene. In fact, Snf1 stimulates the binding of Gcn5 and the acetylation of histone H3 at ADY2 promoter, promoting the transcription of this gene [62].
–
Snf1 and the regulation of metabolism
Besides its role in regulating the transcription of several genes involved in metabolism, Snf1 directly regulates, through phosphorylation, important metabolic enzymes. In fact, together with the class of transcription factors and regulators, proteins linked to metabolism are the most abundant among Snf1 interactors (Fig. 2). Probably the most impactful function exerted by Snf1 as a direct regulator of metabolism is the regulation of the acetyl-CoA carboxylase Acc1 [63]. In yeast, loss of Snf1 causes a dramatic accumulation of fatty acids and the carbon overflow into the fatty acid biosynthetic pathway has been shown to cause inositol auxotrophy mediated by the impairment of INO1 expression [63][64]. Moreover, the excessive allocation of carbon into fatty acids causes a depletion of the intracellular acetyl-CoA pool, and thus a global reduction of acetylation of histones, of Swi4, the DNA-binding protein of the transcription factor SBF [65], and of the β-subunit Sip2 [66].
–
Snf1 was also shown to phosphorylate Pfk27, the second isoform of 6-phosphofructo-2-kinase [67]. Upon glucose removal, Snf1 phosphorylates Pfk27 in its N-terminal domain, leading to the SCFGrr1-dependent degradation of Pfk27 [67]. In particular, Snf1-dependent phosphorylation is required to promote Pfk27 association with the F-box protein Grr1 [67], thus leading to Pfk27 turnover and consequently to a reduction of fructose-2,6-bisphosphate. The importance of Pfk27 turnover is highlighted by the fact that expression of a non-phosphorylatable and non-degradable Pfk27 protein inhibits growth on glycerol [67].
–
Moreover, Snf1 phosphorylates Gpd2, the glycerol-3-phosphate dehydrogenase required for anaerobic growth, thus inhibiting glycerol synthesis during the diauxic shift. In fact, it was reported that Snf1 phosphorylates Gpd2 on Ser72 priming Gpd2 for subsequent phosphorylation on Ser75, probably by Yck1 [68].
–
Snf1 and PKA crosstalk
In yeast, the main pathway activated by glucose is the PKA pathway, involved in metabolism, growth and proliferation [69][70][71]. Targets of PKA include glycolytic and gluconeogenetic enzymes, proteins involved in the metabolism of storage carbohydrates, transcription factors regulating stress response, ribosomal biogenesis, and carbohydrate metabolism. Active PKA directly stimulates glycolysis, cell growth and cell cycle progression, at the same time gluconeogenesis, stress resistance and mobilization of glycogen and trehalose are down-regulated [69][72]. Several examples of cross-talk between Snf1 and PKA pathways have been reported [73]. Indeed, both kinases regulate the activity of the same transcription factors. Adr1, the transcriptional activator of glucose-repressed genes, is inactivated by PKA and activated by Snf1 which promotes its phosphorylation [74][75]. Msn2, the stress-responsive transcriptional activator, which is a well-known target of PKA, is phosphorylated also by Snf1 in glucose starvation [76]. PKA indirectly controls the localization of the β subunit Sip1 and, as a consequence, of the Snf1-Sip1 complex [12]. In addition, PKA contributes to regulate Sak1, one of the Snf1-activating kinases [77].
–
Notably, recent data nicely complement observations of a cross-talk between Snf1 and PKA. Indeed, the adenylate cyclase Cyr1 and Snf1 interact in a nutrient-independent manner [29]. Active Snf1 phosphorylates Cyr1 and negatively regulates cAMP content and PKA-dependent transcription [29]. Moreover, loss of Snf1 causes an alteration in the phosphorylation pattern of adenylate cyclase [29], suggesting that the crosstalk between Snf1 and PKA is more complex than actually reported and needs to be further investigated.
–
Snf1 and the regulation of TORC1
The Target Of Rapamycin Complex I (TORC1) is a highly conserved nutrient-responsive regulator of cell growth and metabolism in all eukaryotes [78][79][80]. Contrary to AMPK, which is active under nutrient-poor conditions, TORC1 is active under nutrient-rich conditions in budding yeast and also in the presence of growth factors in higher eukaryotes [81][82]. In yeast, TORC1 is composed of Tor1, Kog1/Raptor, Lst8 and Tco89 [79][83][84]. Kog1/Raptor is known to recruit substrates such as 4EBP1 and ribosomal S6 kinase (S6K) to the TORC1 complex [85][86] and is required for the regulation of its activity [78][87].
–
Kog1 is phosphorylated by Snf1 under glucose deprivation, as AMPK does on the ortholog Raptor [88], confirming a conserved regulatory function of Snf1/AMPK on TORC1 complex. Nevertheless, the role of Snf1-dependent phosphorylation on Kog1 is somehow different, since Snf1 phosphorylation stimulates the dissociation of the Kog1-Tor1 complex and the formation of Kog1-bodies by limiting the level of active TORC1 complex in the cell [88]. Thus, although the final result of Snf1 phosphorylation is the inactivation of TORC1 activity, this is reached by increasing the activation threshold of TORC1 to guarantee a cellular commitment to a quiescent state and then survival in starvation condition [88].
–
In the presence of nutrients, TORC1 phosphorylates and activates Sch9, the ortholog of S6K in yeast which, together with other substrates, drives ribosome biosynthesis [89][90][91]. Through an in vitro kinase assay and epistasis analysis, Sch9 has been shown to be also a target of Snf1, indeed total Sch9 phosphorylation is reduced in snf1Δ mutant [92]. On the other hand, Snf1-hyperactive cells display a dramatic decrease of TORC1 activity [93]. Moreover, Snf1 activity is required for the downregulation of TORC1-dependent phosphorylation on Sch9 also in glucose deprivation [94].
–
Interestingly, Orlova and coworkers showed that rapamycin treatment results in a significant increase of Thr210 phosphorylation on Snf1 [95], suggesting a reciprocal regulation of Snf1 by TORC1 and a more complex crosstalk between the two signaling pathways.
–
TORC1 activity is involved in the regulation of autophagy, a cellular recycling system that degrades proteins and organelles by delivery to the vacuole in response to nutrient deprivation [96][97]. In yeast, nitrogen starvation, which induces TORC1 inhibition, Atg13 dephosphorylation as well as Atg1 phosphorylation, results in activation of autophagy [98]. Remarkably, although Snf1 has been proposed as a positive regulator of nitrogen-induced autophagy probably because of its phosphorylation on Atg1 [99], in snf1Δ cells, the translocation of GFP-Atg8 to the vacuole is reduced by 50% compared to the wild type [100]. Moreover, Snf1 activity is essential for glucose starvation-induced autophagy, and mitochondrial respiration is a required feature for this energy deprivation condition [100]. These interesting results indicate that further investigations are required to better elucidate the different mechanisms which regulate nitrogen- and glucose-induced autophagy, as well as how Snf1 is involved in such a regulation.
–
Snf1 and the regulation of stress response
Besides nutritional deprivation, Snf1 is also involved in the response to other cellular stresses. Snf1 activity protects against toxicity caused by cadmium [101], hygromycin B [26], hydroxyurea [24], selenite [102], and iron [103]. Snf1 also regulates HSF (Heat Shock transcription Factor) ensuring the cellular resistance to high temperature, oxidative stress [104][105] and counteracts the activity of the transcriptional inhibitor Nrg1, promoting the expression of ENA1, responsible for Na+ ions detoxification [106].
–
In addition, protein kinase Snf1 regulates the Unfolded Protein Response (UPR), the evolutionary conserved pathway activated when improperly folded proteins accumulate and induce endoplasmic reticulum (ER) stress [107][108]. ER misfunction causes severe disease conditions [108], thus the elucidation of the molecular mechanism by which AMPK regulates UPR signaling attracts the increasing interest of cell biologists. Nevertheless, the role of Snf1 in this pathway is still not clear, since partially discrepant data were published on yeast [109][110]. Mizumo and coworkers support a negative role of Snf1 in such a regulation, showing that the deletion of SNF1 gene and Snf1 activation cause increased and decreased resistance to ER stress, respectively [110]. On the contrary, although results from Casamayor’s laboratory highlight that Snf1 activation induces hypersensitivity to ER-stress-inducer agents [109], they also reported that snf1Δ cells are more sensitive to tunicamycin, a known inducer of ER stress [111].
–
Thus, even though these data indicate an interesting role for Snf1 in the regulation of ER stress response, more work is needed to understand the underlying molecular mechanism.
–
Snf1 and the regulation of DNA damage
Interesting results from Simpson-Lavy and collaborators recently show cross-talk between Snf1 and protein kinases involved in DNA damage (Mec1/ATR and Tel1/ATM) [112]. Phosphorylation of the SUMO E3 ligase Mms21 by Mec1 and Tel1 induces SUMOylation and inactivation of Snf1, in response to DNA damage. Thus, fermentation increases while respiration is switched off. Remarkably, inactivation of Snf1 activity by SUMOylation does not affects its phosphorylation at Thr210, indicating that SUMOylation and phosphorylation of Snf1 are independently regulated. The authors suggest that this metabolic switch may protect yeast cells from oxidative stress and propose interesting parallelisms with Warburg effect in cancer cells [112].
–
Snf1 and the regulation of aging
Given the role of AMPK kinase in the regulation of energy homeostasis, it is not surprising the existence of a strong relationship between aging and the AMPK pathway [113][114][115][116][117]. Indeed, hallmarks of the aging process such as mitochondrial dysfunction [118], autophagy [119], endoplasmic reticulum stress [120] and DNA damage repair [121] are regulated by AMPK. Importantly, both pharmacological stimulation and exercise increase AMPK activity in skeletal muscle of young rats but not in old ones [122]. Moreover, AMPK-dependent acetyl-CoA carboxylase and mitochondrial biogenesis are impaired with aging [122]. These and other results suggest that the basal activity of AMPK declines with aging, contributing to the dysregulation of intracellular metabolism.
–
The first evidence of the involvement of Snf1/AMPK in the aging process in yeast occurred several years ago when Asharafhi and co-workers discovered that Snf1 activity increased in replicative aging even in the presence of abundant glucose in the environment [14]. Moreover, loss of the β-subunit Sip2 accelerates aging [14] and Sip2 acetylation enhances its interaction with Snf1, decreases the kinase activity of the complex and extends Replicative Life Span (RLS, an aging model of mitotically active cells) [92]. Thus, in a yeast model of replicative aging, these results indicate that Snf1 inactivation promotes longevity [92].
–
On the contrary, in a yeast model of Chronological Life Span (CLS, a model of aging in post-mitotic mammalian cells), Snf1 activity is critical for the extension of CLS in caloric restriction condition and cells deleted for SNF1 gene have a very short CLS [123][124]. Accordingly, the upregulation of Snf1 activity extends the life span [125] and drugs like metformin (which activates AMPK) are being proposed for the treatment of age-associated disorders [126].
–
Taken together these data indicate that Snf1 activity promotes longevity in CLS while accelerates RLS, showing distinct and opposite mechanisms in the regulation of aging, as also reported for other proteins in yeast [127][128].
NON CONVENTIONAL ROLES OF SNF1
Snf1 and the regulation of endocytosis
Recent data suggest that Snf1 function is not limited to conditions of carbon limitation. One of such functions is the regulation of proteins involved in endocytosis and cellular trafficking, indeed several interactors and substrates of Snf1 are in this cluster (Fig. 2). Snf1 interacts with and phosphorylates the α-arrestin Rod1 [27][129][130], which regulates endocytosis of the lactate transporter Jen1 and of the hexose transporters Hxt1, Hxt3 and Hxt6, in response to glucose. Remarkably, Snf1 phosphorylation on Rod1 occurs not only in glucose limitation, but also in the presence of glucose. In this condition, Snf1 phosphorylation inhibits Rod1-mediated trafficking of Hxt1 and Hxt3, thus maintaining a high glucose transporter activity [131]. Therefore, Snf1-mediated phosphorylation has both an inhibitory and activatory function on the trafficking of hexose transporters, depending on the level of Snf1-mediated phosphorylation on Rod1 [131]. According to the proposed model, when Snf1 activity is high, Rod1 is hyper-phosphorylated and the endocytosis of hexose transporters is active. On the other hand, in glucose growing cells, Snf1 activity is low, Rod1 is hypo-phosphorylated and thus its trafficking function is inhibited, indicating that Snf1 retains physiologically important function also under high-glucose conditions, probably to direct its activity to specific targets [131].
–
Snf1 also regulates Arf3 [132], one of the ADP-ribosylation factors (Arfs), involved in vesicle transport and actin reorganization. Yeast Arf3 is required for invasive growth and its activity is stimulated upon glucose-depletion in a Snf1-dependent manner. The regulation of invasive growth in nutrient depletion is actually a conventional role of Snf1 [133][134]. However, the peculiarity of the activation of Arf3 is that it does not depend on Snf1 kinase activity, but rather on its absolutely new role as a GEF (Guanine Nucleotide Exchange Factor). In fact, it was shown that the C-terminal hydrophobic α-helix core of Snf1 is a non-canonical GEF for Arf3 activation [132].
–
Snf1 and the regulation of cell proliferation, cell cycle and metabolism
Several reports indicate that Snf1 could be active in the presence of high glucose [52][131][135][136], indicating regulatory roles for Snf1 under glucose repression. In facts, cells lacking Snf1 (both snf1Δ and snf1as mutant, whose activity can be chemically inhibited) show a slow growth phenotype and an increased fraction of cells in G1 phase, in synthetic medium supplemented with 2% glucose [25][137]. Nevertheless, no perturbations of growth occur in complete (YPD, with 2% glucose) and in synthetic media with 5% glucose [25][137], as well as in complete synthetic medium (our unpublished results), indicating that the nutritional composition of the media may influence the cellular requirement for Snf1 function.
–
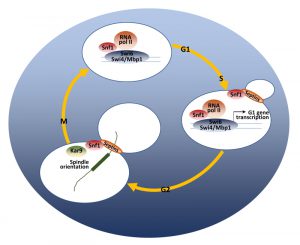
The relevant role of the catalytic activity of Snf1 in regulating proliferation in glucose-repressed condition is further supported by the fact that Snf1 loss reduces the expression of G1-specific genes [137]. The G1/S transition is regulated by the expression of about 150 genes (the G1 regulon) [138], controlled by the transcription factors SBF (Swi4-Swi6) and MBF (Mbp1-Swi6) [65]. Snf1 directs the expression of both SBF and MBF-regulated genes [25][137], by modulating the recruitment of both the co-activator Swi6 and the RNA polymerase II to the promoters of G1-genes [137] (Fig. 3).
–
Snf1-T210 is weakly phosphorylated in 2% glucose, confirming that it is partially active in that growth condition [21][137][139]. Consistently, the non phosphorylatable SNF1-T210A mutant shows a slow growth phenotype and a delayed G1/S transition. [25][137].
–
Snf1 exerts its function in mitosis too and active Snf1 is localized at the division site from the time of bud emergence to cytokinesis [140]. Both septins and protein kinase Elm1 are required for proper Snf1 localization to the bud neck, indicating that the presence of an accurate scaffold is necessary for this process (Fig. 3). Loss of Snf1 activity causes a defect in the correct alignment of the mitotic spindle, that in turn induces a delay of the metaphase-to-anaphase transition, thus clearly indicating Snf1 function for proper spindle orientation. Two major pathways are responsible for the spindle alignment along the mother-bud axis in budding yeast: the Kar9-pathway and the Dyn1-pathway [141]. Recent results show that Snf1 acts in parallel to Dyn1 and in concert with Kar9 to promote spindle positioning, probably phosphorylating components of the Kar9-dependent pathway [140]. In support of these data, several key regulators of cell cycle are known interactors of Snf1, mainly involved in the network of spindle orientation, mitotic exit and cytokinesis (Fig. 2).
–
It is amazing that cells lacking Snf1 and growing in synthetic media containing 2% glucose, show an extensive transcriptional reprogramming, being the most upregulated genes mainly involved in transmembrane transport and metabolic processes such as aminoacid biosynthesis, iron homeostasis and redox metabolism [142]. Moreover, an increase of cellular dependence on mitochondrial function in glucose repression condition is clearly noticeable in snf1Δ cells, further supporting the emerging roles of Snf1 in non-limiting nutrient condition too [142].
PERSPECTIVES AND CONCLUDING REMARKS
Many studies have reported that AMPK activity is altered in several diseases, such as inflammation, diabetes and cancer [143][144]. Moreover, the number of pharmacological agents that activate AMPK has continued to increase and some of them are promise hypoglycemic agents. Importantly, although AMPK is considered a key target for cancer treatment, emerging data indicate that AMPK performs both anti- and pro-tumorigenic roles depending on the composition of AMPK complex, signaling networks and environmental conditions [145][146]. The pro-tumorigenic role of AMPK involves promotion of metabolic adaptation for cancer cell survival by regulating fatty acid metabolism and maintaining the ability to growth in stressful conditions [143][147].
–
Therefore, the expansion of the repertoire of AMPK substrates, as well as more in-depth studies of the molecular mechanisms by which AMPK is activated, will help to better understand the roles of this kinase in the regulation of human pathologies. In this context, the yeast unicellular organism Saccharomyces cerevisiae is a powerful model for studying fundamental aspects of eukaryotic cell biology and to validate the increasing downstream targets of the class of Snf1/AMPK protein kinases which control the complexity of cellular physiology.
References
- S. Herzig, and R.J. Shaw, "AMPK: guardian of metabolism and mitochondrial homeostasis", Nature Reviews Molecular Cell Biology, vol. 19, pp. 121-135, 2017. http://dx.doi.org/10.1038/nrm.2017.95
- R. Ghillebert, E. Swinnen, J. Wen, L. Vandesteene, M. Ramon, K. Norga, F. Rolland, and J. Winderickx, "The AMPK/SNF1/SnRK1 fuel gauge and energy regulator: structure, function and regulation", The FEBS Journal, vol. 278, pp. 3978-3990, 2011. http://dx.doi.org/10.1111/j.1742-4658.2011.08315.x
- K. Hedbacker, "SNF1/AMPK pathways in yeast", Frontiers in Bioscience, vol. 13, pp. 2408, 2008. http://dx.doi.org/10.2741/2854
- M. Carlson, B.C. Osmond, and D. Botstein, "Mutants of yeast defective in sucrose utilization.", Genetics, 1981. http://www.ncbi.nlm.nih.gov/pubmed/7040163
- M.J. Rudolph, G.A. Amodeo, Y. Bai, and L. Tong, "Crystal structure of the protein kinase domain of yeast AMP-activated protein kinase Snf1", Biochemical and Biophysical Research Communications, vol. 337, pp. 1224-1228, 2005. http://dx.doi.org/10.1016/j.bbrc.2005.09.181
- L. Chen, Z. Jiao, L. Zheng, Y. Zhang, S. Xie, Z. Wang, and J. Wu, "Structural insight into the autoinhibition mechanism of AMP-activated protein kinase", Nature, vol. 459, pp. 1146-1149, 2009. http://dx.doi.org/10.1038/nature08075
- M.C. Schmidt, "beta-subunits of Snf1 kinase are required for kinase function and substrate definition", The EMBO Journal, vol. 19, pp. 4936-4943, 2000. http://dx.doi.org/10.1093/emboj/19.18.4936
- J.R. Erickson, and M. Johnston, "Genetic and molecular characterization of GAL83: its interaction and similarities with other genes involved in glucose repression in Saccharomyces cerevisiae.", Genetics, 1993. http://www.ncbi.nlm.nih.gov/pubmed/8293971
- X. Yang, R. Jiang, and M. Carlson, "A family of proteins containing a conserved domain that mediates interaction with the yeast SNF1 protein kinase complex.", The EMBO journal, 1994. http://www.ncbi.nlm.nih.gov/pubmed/7813428
- R. Jiang, and M. Carlson, "The Snf1 Protein Kinase and Its Activating Subunit, Snf4, Interact with Distinct Domains of the Sip1/Sip2/Gal83 Component in the Kinase Complex", Molecular and Cellular Biology, vol. 17, pp. 2099-2106, 1997. http://dx.doi.org/10.1128/MCB.17.4.2099
- O. Vincent, R. Townley, S. Kuchin, and M. Carlson, "Subcellular localization of the Snf1 kinase is regulated by specific β subunits and a novel glucose signaling mechanism", Genes & Development, vol. 15, pp. 1104-1114, 2001. http://dx.doi.org/10.1101/gad.879301
- K. Hedbacker, R. Townley, and M. Carlson, "Cyclic AMP-dependent protein kinase regulates the subcellular localization of Snf1-Sip1 protein kinase.", Molecular and cellular biology, 2004. http://www.ncbi.nlm.nih.gov/pubmed/14966266
- N. Nath, R.R. McCartney, and M.C. Schmidt, "Purification and Characterization of Snf1 Kinase Complexes Containing a Defined β Subunit Composition", Journal of Biological Chemistry, vol. 277, pp. 50403-50408, 2002. http://dx.doi.org/10.1074/jbc.M207058200
- K. Ashrafi, S.S. Lin, J.K. Manchester, and J.I. Gordon, "Sip2p and its partner snf1p kinase affect aging in S. cerevisiae.", Genes & development, 2000. http://www.ncbi.nlm.nih.gov/pubmed/10921902
- K. Hedbacker, and M. Carlson, "Regulation of the Nucleocytoplasmic Distribution of Snf1-Gal83 Protein Kinase", Eukaryotic Cell, vol. 5, pp. 1950-1956, 2006. http://dx.doi.org/10.1128/EC.00256-06
- O. Vincent, "Gal83 mediates the interaction of the Snf1 kinase complex with the transcription activator Sip4", The EMBO Journal, vol. 18, pp. 6672-6681, 1999. http://dx.doi.org/10.1093/emboj/18.23.6672
- S. Kuchin, I. Treich, and M. Carlson, "A regulatory shortcut between the Snf1 protein kinase and RNA polymerase II holoenzyme", Proceedings of the National Academy of Sciences, vol. 97, pp. 7916-7920, 2000. http://dx.doi.org/10.1073/pnas.140109897
- M. Momcilovic, S.H. Iram, Y. Liu, and M. Carlson, "Roles of the Glycogen-binding Domain and Snf4 in Glucose Inhibition of SNF1 Protein Kinase", Journal of Biological Chemistry, vol. 283, pp. 19521-19529, 2008. http://dx.doi.org/10.1074/jbc.M803624200
- J.L. Celenza, and M. Carlson, "Mutational analysis of the Saccharomyces cerevisiae SNF1 protein kinase and evidence for functional interaction with the SNF4 protein.", Molecular and Cellular Biology, vol. 9, pp. 5034-5044, 1989. http://dx.doi.org/10.1128/MCB.9.11.5034
- A. Leech, N. Nath, R.R. McCartney, and M.C. Schmidt, "Isolation of Mutations in the Catalytic Domain of the Snf1 Kinase That Render Its Activity Independent of the Snf4 Subunit", Eukaryotic Cell, vol. 2, pp. 265-273, 2003. http://dx.doi.org/10.1128/EC.2.2.265-273.2003
- R.R. McCartney, and M.C. Schmidt, "Regulation of Snf1 Kinase", Journal of Biological Chemistry, vol. 276, pp. 36460-36466, 2001. http://dx.doi.org/10.1074/jbc.M104418200
- S. Hong, F.C. Leiper, A. Woods, D. Carling, and M. Carlson, "Activation of yeast Snf1 and mammalian AMP-activated protein kinase by upstream kinases", Proceedings of the National Academy of Sciences, vol. 100, pp. 8839-8843, 2003. http://dx.doi.org/10.1073/pnas.1533136100
- C.M. Sutherland, S.A. Hawley, R.R. McCartney, A. Leech, M.J. Stark, M.C. Schmidt, and D. Hardie, "Elm1p Is One of Three Upstream Kinases for the Saccharomyces cerevisiae SNF1 Complex", Current Biology, vol. 13, pp. 1299-1305, 2003. http://dx.doi.org/10.1016/S0960-9822(03)00459-7
- C. Dubacq, A. Chevalier, and C. Mann, "The Protein Kinase Snf1 Is Required for Tolerance to the Ribonucleotide Reductase Inhibitor Hydroxyurea", Molecular and Cellular Biology, vol. 24, pp. 2560-2572, 2004. http://dx.doi.org/10.1128/MCB.24.6.2560-2572.2004
- S. Pessina, V. Tsiarentsyeva, S. Busnelli, M. Vanoni, L. Alberghina, and P. Coccetti, "Snf1/AMPK promotes S-phase entrance by controllingCLB5transcription in budding yeast", Cell Cycle, vol. 9, pp. 2189-2200, 2010. http://dx.doi.org/10.4161/cc.9.11.11847
- F. Portillo, J.M. Mulet, and R. Serrano, "A role for the non‐phosphorylated form of yeast Snf1: tolerance to toxic cations and activation of potassium transport", FEBS Letters, vol. 579, pp. 512-516, 2004. http://dx.doi.org/10.1016/j.febslet.2004.12.019
- J. Shinoda, and Y. Kikuchi, "Rod1, an arrestin-related protein, is phosphorylated by Snf1-kinase in Saccharomyces cerevisiae", Biochemical and Biophysical Research Communications, vol. 364, pp. 258-263, 2007. http://dx.doi.org/10.1016/j.bbrc.2007.09.134
- S.L. Wade, K. Poorey, S. Bekiranov, and D.T. Auble, "The Snf1 kinase and proteasome-associated Rad23 regulate UV-responsive gene expression", The EMBO Journal, vol. 28, pp. 2919-2931, 2009. http://dx.doi.org/10.1038/emboj.2009.229
- R. Nicastro, F. Tripodi, M. Gaggini, A. Castoldi, V. Reghellin, S. Nonnis, G. Tedeschi, and P. Coccetti, "Snf1 Phosphorylates Adenylate Cyclase and Negatively Regulates Protein Kinase A-dependent Transcription in Saccharomyces cerevisiae", Journal of Biological Chemistry, vol. 290, pp. 24715-24726, 2015. http://dx.doi.org/10.1074/jbc.M115.658005
- K. Ludin, R. Jiang, and M. Carlson, "Glucose-regulated interaction of a regulatory subunit of protein phosphatase 1 with the Snf1 protein kinase in Saccharomyces cerevisiae", Proceedings of the National Academy of Sciences, vol. 95, pp. 6245-6250, 1998. http://dx.doi.org/10.1073/pnas.95.11.6245
- P. Sanz, G.R. Alms, T.A.J. Haystead, and M. Carlson, "Regulatory Interactions between the Reg1-Glc7 Protein Phosphatase and the Snf1 Protein Kinase", Molecular and Cellular Biology, vol. 20, pp. 1321-1328, 2000. http://dx.doi.org/10.1128/MCB.20.4.1321-1328.2000
- D.L. Frederick, and K. Tatchell, "The REG2 Gene of Saccharomyces cerevisiae Encodes a Type 1 Protein Phosphatase-Binding Protein That Functions with Reg1p and the Snf1 Protein Kinase to Regulate Growth", Molecular and Cellular Biology, vol. 16, pp. 2922-2931, 1996. http://dx.doi.org/10.1128/MCB.16.6.2922
- D. Huang, I. Farkas, and P.J. Roach, "Pho85p, a Cyclin-Dependent Protein Kinase, and the Snf1p Protein Kinase Act Antagonistically To Control Glycogen Accumulation in Saccharomyces cerevisiae", Molecular and Cellular Biology, vol. 16, pp. 4357-4365, 1996. http://dx.doi.org/10.1128/MCB.16.8.4357
- E.M. Rubenstein, R.R. McCartney, C. Zhang, K.M. Shokat, M.K. Shirra, K.M. Arndt, and M.C. Schmidt, "Access Denied: Snf1 Activation Loop Phosphorylation Is Controlled by Availability of the Phosphorylated Threonine 210 to the PP1 Phosphatase", Journal of Biological Chemistry, vol. 283, pp. 222-230, 2008. http://dx.doi.org/10.1074/jbc.M707957200
- S. Dale, W.A. Wilson, A.M. Edelman, and D. Hardie, "Similar substrate recognition motifs for mammalian AMP‐activated protein kinase, higher plant HMG‐CoA reductase kinase‐A, yeast SNF1, and mammalian calmodulin‐dependent protein kinase I", FEBS Letters, vol. 361, pp. 191-195, 1995. http://dx.doi.org/10.1016/0014-5793(95)00172-6
- W.A. Wilson, S.A. Hawley, and D. Hardie, "Glucose repression/derepression in budding yeast: SNF1 protein kinase is activated by phosphorylation under derepressing conditions, and this correlates with a high AMP:ATP ratio", Current Biology, vol. 6, pp. 1426-1434, 1996. http://dx.doi.org/10.1016/S0960-9822(96)00747-6
- F. Mayer, R. Heath, E. Underwood, M. Sanders, D. Carmena, R. McCartney, F. Leiper, B. Xiao, C. Jing, P. Walker, L. Haire, R. Ogrodowicz, S. Martin, M. Schmidt, S. Gamblin, and D. Carling, "ADP Regulates SNF1, the Saccharomyces cerevisiae Homolog of AMP-Activated Protein Kinase", Cell Metabolism, vol. 14, pp. 707-714, 2011. http://dx.doi.org/10.1016/j.cmet.2011.09.009
- D.G. Chandrashekarappa, R.R. McCartney, and M.C. Schmidt, "Subunit and Domain Requirements for Adenylate-mediated Protection of Snf1 Kinase Activation Loop from Dephosphorylation*", Journal of Biological Chemistry, vol. 286, pp. 44532-44541, 2011. http://dx.doi.org/10.1074/jbc.M111.315895
- V. Nayak, K. Zhao, A. Wyce, M.F. Schwartz, W. Lo, S.L. Berger, and R. Marmorstein, "Structure and Dimerization of the Kinase Domain from Yeast Snf1, a Member of the Snf1/AMPK Protein Family", Structure, vol. 14, pp. 477-485, 2006. http://dx.doi.org/10.1016/j.str.2005.12.008
- R.R. McCartney, L. Garnar-Wortzel, D.G. Chandrashekarappa, and M.C. Schmidt, "Activation and inhibition of Snf1 kinase activity by phosphorylation within the activation loop", Biochimica et Biophysica Acta (BBA) - Proteins and Proteomics, vol. 1864, pp. 1518-1528, 2016. http://dx.doi.org/10.1016/j.bbapap.2016.08.007
- K.J. Simpson-Lavy, and M. Johnston, "SUMOylation regulates the SNF1 protein kinase", Proceedings of the National Academy of Sciences, vol. 110, pp. 17432-17437, 2013. http://dx.doi.org/10.1073/pnas.1304839110
- M.A. Wilson, E. Koutelou, C. Hirsch, K. Akdemir, A. Schibler, M.C. Barton, and S.Y.R. Dent, "Ubp8 and SAGA Regulate Snf1 AMP Kinase Activity", Molecular and Cellular Biology, vol. 31, pp. 3126-3135, 2011. http://dx.doi.org/10.1128/MCB.01350-10
- R. Jiao, S. Postnikoff, T.A. Harkness, and T.G. Arnason, "The SNF1 Kinase Ubiquitin-associated Domain Restrains Its Activation, Activity, and the Yeast Life Span", Journal of Biological Chemistry, vol. 290, pp. 15393-15404, 2015. http://dx.doi.org/10.1074/jbc.M115.647032
- E.T. Young, C. Zhang, K.M. Shokat, P.K. Parua, and K.A. Braun, "The AMP-activated Protein Kinase Snf1 Regulates Transcription Factor Binding, RNA Polymerase II Activity, and mRNA Stability of Glucose-repressed Genes in Saccharomyces cerevisiae", Journal of Biological Chemistry, vol. 287, pp. 29021-29034, 2012. http://dx.doi.org/10.1074/jbc.M112.380147
- D. Hedges, M. Proft, and K. Entian, "CAT8, a New Zinc Cluster-Encoding Gene Necessary for Derepression of Gluconeogenic Enzymes in the Yeast Saccharomyces cerevisiae", Molecular and Cellular Biology, vol. 15, pp. 1915-1922, 1995. http://dx.doi.org/10.1128/MCB.15.4.1915
- M.A. Treitel, S. Kuchin, and M. Carlson, "Snf1 Protein Kinase Regulates Phosphorylation of the Mig1 Repressor in Saccharomyces cerevisiae", Molecular and Cellular Biology, vol. 18, pp. 6273-6280, 1998. http://dx.doi.org/10.1128/MCB.18.11.6273
- P. Lesage, X. Yang, and M. Carlson, "Yeast SNF1 Protein Kinase Interacts with SIP4, a C6 Zinc Cluster Transcriptional Activator: a New Role for SNF1 in the Glucose Response", Molecular and Cellular Biology, vol. 16, pp. 1921-1928, 1996. http://dx.doi.org/10.1128/MCB.16.5.1921
- W. Lo, L. Duggan, N.C. Tolga, . Emre, R. Belotserkovskya, W.S. Lane, R. Shiekhattar, and S.L. Berger, "Snf1--a Histone Kinase That Works in Concert with the Histone Acetyltransferase Gcn5 to Regulate Transcription", Science, vol. 293, pp. 1142-1146, 2001. http://dx.doi.org/10.1126/science.1062322
- J.O. Nehlin, and H. Ronne, "Yeast MIG1 repressor is related to the mammalian early growth response and Wilms' tumour finger proteins.", The EMBO journal, 1990. http://www.ncbi.nlm.nih.gov/pubmed/2167835
- M.J. DeVit, and M. Johnston, "The nuclear exportin Msn5 is required for nuclear export of the Mig1 glucose repressor of Saccharomyces cerevisiae", Current Biology, vol. 9, pp. 1231-1241, 1999. http://dx.doi.org/10.1016/S0960-9822(99)80503-X
- M. Papamichos‐Chronakis, T. Gligoris, and D. Tzamarias, "The Snf1 kinase controls glucose repression in yeast by modulating interactions between the Mig1 repressor and the Cyc8‐Tup1 co‐repressor", EMBO reports, vol. 5, pp. 368-372, 2004. http://dx.doi.org/10.1038/sj.embor.7400120
- D. Ahuatzi, A. Riera, R. Pela´ez, P. Herrero, and F. Moreno, "Hxk2 Regulates the Phosphorylation State of Mig1 and Therefore Its Nucleocytoplasmic Distribution", Journal of Biological Chemistry, vol. 282, pp. 4485-4493, 2007. http://dx.doi.org/10.1074/jbc.M606854200
- S. Ostergaard, K.O. Walløe, C.S. Gomes, L. Olsson, and J. Nielsen, "The impact ofGAL6,GAL80, andMIG1on glucose control of theGALsystem inSaccharomyces cerevisiae", FEMS Yeast Research, vol. 1, pp. 47-55, 2001. http://dx.doi.org/10.1111/j.1567-1364.2001.tb00012.x
- C.J. van Oevelen, H.A. van Teeffelen, F.J. van Werven, and H.T.M. Timmers, "Snf1p-dependent Spt-Ada-Gcn5-acetyltransferase (SAGA) Recruitment and Chromatin Remodeling Activities on the HXT2 and HXT4 Promoters", Journal of Biological Chemistry, vol. 281, pp. 4523-4531, 2006. http://dx.doi.org/10.1074/jbc.M509330200
- S. Hohmann, P. Van Dijck, K. Luyten, and J.M. Thevelein, "The byp1-3 allele of the Saccharomyces cerevisiae GGS1/TPS1 gene and its multi-copy suppressor tRNA(GLN) (CAG): Ggs1/Tps1 protein levels restraining growth on fermentable sugars and trehalose accumulation.", Current genetics, 1994. http://www.ncbi.nlm.nih.gov/pubmed/7882422
- C. Jin, A. Barrientos, C.B. Epstein, R.A. Butow, and A. Tzagoloff, "SIT4 regulation of Mig1p‐mediated catabolite repression in Saccharomyces cerevisiae", FEBS Letters, vol. 581, pp. 5658-5663, 2007. http://dx.doi.org/10.1016/j.febslet.2007.11.027
- O. Vincent, and M. Carlson, "Sip4, a Snf1 kinase-dependent transcriptional activator, binds to the carbon source-responsive element of gluconeogenic genes", The EMBO Journal, vol. 17, pp. 7002-7008, 1998. http://dx.doi.org/10.1093/emboj/17.23.7002
- F. Randez-Gil, N. Bojunga, M. Proft, and K. Entian, "Glucose Derepression of Gluconeogenic Enzymes in Saccharomyces cerevisiae Correlates with Phosphorylation of the Gene Activator Cat8p", Molecular and Cellular Biology, vol. 17, pp. 2502-2510, 1997. http://dx.doi.org/10.1128/MCB.17.5.2502
- E.T. Young, N. Kacherovsky, and K. Van Riper, "Snf1 Protein Kinase Regulates Adr1 Binding to Chromatin but Not Transcription Activation", Journal of Biological Chemistry, vol. 277, pp. 38095-38103, 2002. http://dx.doi.org/10.1074/jbc.M206158200
- N. Kacherovsky, C. Tachibana, E. Amos, D. Fox, and E.T. Young, "Promoter Binding by the Adr1 Transcriptional Activator May Be Regulated by Phosphorylation in the DNA-Binding Region", PLoS ONE, vol. 3, pp. e3213, 2008. http://dx.doi.org/10.1371/journal.pone.0003213
- M.K. Shirra, R.R. McCartney, C. Zhang, K.M. Shokat, M.C. Schmidt, and K.M. Arndt, "A Chemical Genomics Study Identifies Snf1 as a Repressor of GCN4 Translation", Journal of Biological Chemistry, vol. 283, pp. 35889-35898, 2008. http://dx.doi.org/10.1074/jbc.M805325200
- G. Abate, E. Bastonini, K.A. Braun, L. Verdone, E.T. Young, and M. Caserta, "Snf1/AMPK regulates Gcn5 occupancy, H3 acetylation and chromatin remodelling at S. cerevisiae ADY2 promoter", Biochimica et Biophysica Acta (BBA) - Gene Regulatory Mechanisms, vol. 1819, pp. 419-427, 2012. http://dx.doi.org/10.1016/j.bbagrm.2012.01.009
- M.K. Shirra, J. Patton-Vogt, A. Ulrich, O. Liuta-Tehlivets, S.D. Kohlwein, S.A. Henry, and K.M. Arndt, "Inhibition of Acetyl Coenzyme A Carboxylase Activity Restores Expression of the INO1 Gene in a snf1Mutant Strain of Saccharomyces cerevisiae", Molecular and Cellular Biology, vol. 21, pp. 5710-5722, 2001. http://dx.doi.org/10.1128/MCB.21.17.5710-5722.2001
- H. Hofbauer, F. Schopf, H. Schleifer, O. Knittelfelder, B. Pieber, G. Rechberger, H. Wolinski, M. Gaspar, C. Kappe, J. Stadlmann, K. Mechtler, A. Zenz, K. Lohner, O. Tehlivets, S. Henry, and S. Kohlwein, "Regulation of Gene Expression through a Transcriptional Repressor that Senses Acyl-Chain Length in Membrane Phospholipids", Developmental Cell, vol. 29, pp. 729-739, 2014. http://dx.doi.org/10.1016/j.devcel.2014.04.025
- C. Koch, T. Moll, M. Neuberg, H. Ahorn, and K. Nasmyth, "A Role for the Transcription Factors Mbp1 and Swi4 in Progression from G1 to S Phase", Science, vol. 261, pp. 1551-1557, 1993. http://dx.doi.org/10.1126/science.8372350
- M. Zhang, L. Galdieri, and A. Vancura, "The Yeast AMPK Homolog SNF1 Regulates Acetyl Coenzyme A Homeostasis and Histone Acetylation", Molecular and Cellular Biology, vol. 33, pp. 4701-4717, 2013. http://dx.doi.org/10.1128/MCB.00198-13
- J.A. Benanti, S.K. Cheung, M.C. Brady, and D.P. Toczyski, "A proteomic screen reveals SCFGrr1 targets that regulate the glycolytic–gluconeogenic switch", Nature Cell Biology, vol. 9, pp. 1184-1191, 2007. http://dx.doi.org/10.1038/ncb1639
- Y.J. Lee, G.R. Jeschke, F.M. Roelants, J. Thorner, and B.E. Turk, "Reciprocal Phosphorylation of Yeast Glycerol-3-Phosphate Dehydrogenases in Adaptation to Distinct Types of Stress", Molecular and Cellular Biology, vol. 32, pp. 4705-4717, 2012. http://dx.doi.org/10.1128/MCB.00897-12
- M. Conrad, J. Schothorst, H.N. Kankipati, G. Van Zeebroeck, M. Rubio-Texeira, and J.M. Thevelein, "Nutrient sensing and signaling in the yeastSaccharomyces cerevisiae", FEMS Microbiology Reviews, vol. 38, pp. 254-299, 2014. http://dx.doi.org/10.1111/1574-6976.12065
- M. Deprez, E. Eskes, T. Wilms, P. Ludovico, and J. Winderickx, "pH homeostasis links the nutrient sensing PKA/TORC1/Sch9 ménage-à-trois to stress tolerance and longevity", Microbial Cell, vol. 5, pp. 119-136, 2018. http://dx.doi.org/10.15698/mic2018.03.618
- S. Busti, P. Coccetti, L. Alberghina, and M. Vanoni, "Glucose Signaling-Mediated Coordination of Cell Growth and Cell Cycle in Saccharomyces Cerevisiae", Sensors, vol. 10, pp. 6195-6240, 2010. http://dx.doi.org/10.3390/s100606195
- S. Zaman, S.I. Lippman, X. Zhao, and J.R. Broach, "HowSaccharomycesResponds to Nutrients", Annual Review of Genetics, vol. 42, pp. 27-81, 2008. http://dx.doi.org/10.1146/annurev.genet.41.110306.130206
- S. Shashkova, N. Welkenhuysen, and S. Hohmann, "Molecular communication: crosstalk between the Snf1 and other signaling pathways", FEMS Yeast Research, vol. 15, 2015. http://dx.doi.org/10.1093/femsyr/fov026
- J.R. Cherry, T.R. Johnson, C. Dollard, J.R. Shuster, and C.L. Denis, "Cyclic AMP-dependent protein kinase phosphorylates and inactivates the yeast transcriptional activator ADR1", Cell, vol. 56, pp. 409-419, 1989. http://dx.doi.org/10.1016/0092-8674(89)90244-4
- S. Ratnakumar, N. Kacherovsky, E. Arms, and E.T. Young, "Snf1 Controls the Activity of Adr1 Through Dephosphorylation of Ser230", Genetics, vol. 182, pp. 735-745, 2009. http://dx.doi.org/10.1534/genetics.109.103432
- V. De Wever, W. Reiter, A. Ballarini, G. Ammerer, and C. Brocard, "A dual role for PP1 in shaping the Msn2-dependent transcriptional response to glucose starvation", The EMBO Journal, vol. 24, pp. 4115-4123, 2005. http://dx.doi.org/10.1038/sj.emboj.7600871
- L. Barrett, M. Orlova, M. Maziarz, and S. Kuchin, "Protein Kinase A Contributes to the Negative Control of Snf1 Protein Kinase in Saccharomyces cerevisiae", Eukaryotic Cell, vol. 11, pp. 119-128, 2012. http://dx.doi.org/10.1128/EC.05061-11
- D. Kim, D.D. Sarbassov, S.M. Ali, J.E. King, R.R. Latek, H. Erdjument-Bromage, P. Tempst, and D.M. Sabatini, "mTOR Interacts with Raptor to Form a Nutrient-Sensitive Complex that Signals to the Cell Growth Machinery", Cell, vol. 110, pp. 163-175, 2002. http://dx.doi.org/10.1016/S0092-8674(02)00808-5
- R. Loewith, E. Jacinto, S. Wullschleger, A. Lorberg, J.L. Crespo, D. Bonenfant, W. Oppliger, P. Jenoe, and M.N. Hall, "Two TOR Complexes, Only One of which Is Rapamycin Sensitive, Have Distinct Roles in Cell Growth Control", Molecular Cell, vol. 10, pp. 457-468, 2002. http://dx.doi.org/10.1016/S1097-2765(02)00636-6
- S. Wullschleger, R. Loewith, and M.N. Hall, "TOR Signaling in Growth and Metabolism", Cell, vol. 124, pp. 471-484, 2006. http://dx.doi.org/10.1016/j.cell.2006.01.016
- D.M. Sabatini, "mTOR and cancer: insights into a complex relationship", Nature Reviews Cancer, vol. 6, pp. 729-734, 2006. http://dx.doi.org/10.1038/nrc1974
-
A. González, and M.N. Hall, "Nutrient sensing and
TOR signaling in yeast and mammals", The EMBO Journal, vol. 36, pp. 397-408, 2017. http://dx.doi.org/10.15252/embj.201696010 - A. Reinke, S. Anderson, J.M. McCaffery, J. Yates, S. Aronova, S. Chu, S. Fairclough, C. Iverson, K.P. Wedaman, and T. Powers, "TOR Complex 1 Includes a Novel Component, Tco89p (YPL180w), and Cooperates with Ssd1p to Maintain Cellular Integrity in Saccharomyces cerevisiae", Journal of Biological Chemistry, vol. 279, pp. 14752-14762, 2004. http://dx.doi.org/10.1074/jbc.M313062200
- K.P. Wedaman, A. Reinke, S. Anderson, J. Yates, J.M. McCaffery, and T. Powers, "Tor Kinases Are in Distinct Membrane-associated Protein Complexes inSaccharomyces cerevisiae", Molecular Biology of the Cell, vol. 14, pp. 1204-1220, 2003. http://dx.doi.org/10.1091/mbc.E02-09-0609
- H. Nojima, C. Tokunaga, S. Eguchi, N. Oshiro, S. Hidayat, K. Yoshino, K. Hara, N. Tanaka, J. Avruch, and K. Yonezawa, "The Mammalian Target of Rapamycin (mTOR) Partner, Raptor, Binds the mTOR Substrates p70 S6 Kinase and 4E-BP1 through Their TOR Signaling (TOS) Motif", Journal of Biological Chemistry, vol. 278, pp. 15461-15464, 2003. http://dx.doi.org/10.1074/jbc.C200665200
- S.S. Schalm, D.C. Fingar, D.M. Sabatini, and J. Blenis, "TOS Motif-Mediated Raptor Binding Regulates 4E-BP1 Multisite Phosphorylation and Function", Current Biology, vol. 13, pp. 797-806, 2003. http://dx.doi.org/10.1016/S0960-9822(03)00329-4
- K. Hara, Y. Maruki, X. Long, K. Yoshino, N. Oshiro, S. Hidayat, C. Tokunaga, J. Avruch, and K. Yonezawa, "Raptor, a Binding Partner of Target of Rapamycin (TOR), Mediates TOR Action", Cell, vol. 110, pp. 177-189, 2002. http://dx.doi.org/10.1016/S0092-8674(02)00833-4
- J.E. Hughes Hallett, X. Luo, and A.P. Capaldi, "Snf1/AMPK promotes the formation of Kog1/Raptor-bodies to increase the activation threshold of TORC1 in budding yeast", eLife, vol. 4, 2015. http://dx.doi.org/10.7554/eLife.09181
- J. Urban, A. Soulard, A. Huber, S. Lippman, D. Mukhopadhyay, O. Deloche, V. Wanke, D. Anrather, G. Ammerer, H. Riezman, J.R. Broach, C. De Virgilio, M.N. Hall, and R. Loewith, "Sch9 Is a Major Target of TORC1 in Saccharomyces cerevisiae", Molecular Cell, vol. 26, pp. 663-674, 2007. http://dx.doi.org/10.1016/j.molcel.2007.04.020
- A. Huber, S.L. French, H. Tekotte, S. Yerlikaya, M. Stahl, M.P. Perepelkina, M. Tyers, J. Rougemont, A.L. Beyer, and R. Loewith, "Sch9 regulates ribosome biogenesis via Stb3, Dot6 and Tod6 and the histone deacetylase complex RPD3L", The EMBO Journal, vol. 30, pp. 3052-3064, 2011. http://dx.doi.org/10.1038/emboj.2011.221
- P. Jorgensen, I. Rupeš, J.R. Sharom, L. Schneper, J.R. Broach, and M. Tyers, "A dynamic transcriptional network communicates growth potential to ribosome synthesis and critical cell size", Genes & Development, vol. 18, pp. 2491-2505, 2004. http://dx.doi.org/10.1101/gad.1228804
- J. Lu, Y. Lin, J. Sheu, J. Wu, F. Lee, Y. Chen, M. Lin, F. Chiang, T. Tai, S. Berger, Y. Zhao, K. Tsai, H. Zhu, L. Chuang, and J. Boeke, "Acetylation of Yeast AMPK Controls Intrinsic Aging Independently of Caloric Restriction", Cell, vol. 146, pp. 969-979, 2011. http://dx.doi.org/10.1016/j.cell.2011.07.044
- D. DeMille, B.D. Badal, J.B. Evans, A.D. Mathis, J.F. Anderson, and J.H. Grose, "PAS kinase is activated by direct SNF1-dependent phosphorylation and mediates inhibition of TORC1 through the phosphorylation and activation of Pbp1", Molecular Biology of the Cell, vol. 26, pp. 569-582, 2015. http://dx.doi.org/10.1091/mbc.e14-06-1088
- J.E. Hughes Hallett, X. Luo, and A.P. Capaldi, "State Transitions in the TORC1 Signaling Pathway and Information Processing in Saccharomyces cerevisiae", Genetics, vol. 198, pp. 773-786, 2014. http://dx.doi.org/10.1534/genetics.114.168369
- M. Orlova, E. Kanter, D. Krakovich, and S. Kuchin, "Nitrogen Availability and TOR Regulate the Snf1 Protein Kinase in Saccharomyces cerevisiae", Eukaryotic Cell, vol. 5, pp. 1831-1837, 2006. http://dx.doi.org/10.1128/EC.00110-06
- H. Nakatogawa, K. Suzuki, Y. Kamada, and Y. Ohsumi, "Dynamics and diversity in autophagy mechanisms: lessons from yeast", Nature Reviews Molecular Cell Biology, vol. 10, pp. 458-467, 2009. http://dx.doi.org/10.1038/nrm2708
- F. Reggiori, and D.J. Klionsky, "Autophagic Processes in Yeast: Mechanism, Machinery and Regulation", Genetics, vol. 194, pp. 341-361, 2013. http://dx.doi.org/10.1534/genetics.112.149013
- Y. Kamada, "Prime-numbered Atg proteins act at the primary step in autophagy, Unphosphorylatable Atg13 can induce autophagy without TOR inactivation", Autophagy, vol. 6, pp. 415-416, 2010. http://dx.doi.org/10.4161/auto.6.3.11390
- Z. Wang, W.A. Wilson, M.A. Fujino, and P.J. Roach, "Antagonistic Controls of Autophagy and Glycogen Accumulation by Snf1p, the Yeast Homolog of AMP-Activated Protein Kinase, and the Cyclin-Dependent Kinase Pho85p", Molecular and Cellular Biology, vol. 21, pp. 5742-5752, 2001. http://dx.doi.org/10.1128/MCB.21.17.5742-5752.2001
- C. Yi, J. Tong, P. Lu, Y. Wang, J. Zhang, C. Sun, K. Yuan, R. Xue, B. Zou, N. Li, S. Xiao, C. Dai, Y. Huang, L. Xu, L. Li, S. Chen, D. Miao, H. Deng, H. Li, and L. Yu, "Formation of a Snf1-Mec1-Atg1 Module on Mitochondria Governs Energy Deprivation-Induced Autophagy by Regulating Mitochondrial Respiration", Developmental Cell, vol. 41, pp. 59-71.e4, 2017. http://dx.doi.org/10.1016/j.devcel.2017.03.007
- M. Thorsen, G.G. Perrone, E. Kristiansson, M. Traini, T. Ye, I.W. Dawes, O. Nerman, and M.J. Tamás, "Genetic basis of arsenite and cadmium tolerance in Saccharomyces cerevisiae", BMC Genomics, vol. 10, 2009. http://dx.doi.org/10.1186/1471-2164-10-105
- M. Pérez-Sampietro, C. Casas, and E. Herrero, "The AMPK Family Member Snf1 Protects Saccharomyces cerevisiae Cells upon Glutathione Oxidation", PLoS ONE, vol. 8, pp. e58283, 2013. http://dx.doi.org/10.1371/journal.pone.0058283
- L. Li, J. Kaplan, and D.M. Ward, "The glucose sensor Snf1 and the transcription factors Msn2 and Msn4 regulate transcription of the vacuolar iron importer gene CCC1 and iron resistance in yeast", Journal of Biological Chemistry, vol. 292, pp. 15577-15586, 2017. http://dx.doi.org/10.1074/jbc.M117.802504
- S. Hong, and M. Carlson, "Regulation of Snf1 Protein Kinase in Response to Environmental Stress", Journal of Biological Chemistry, vol. 282, pp. 16838-16845, 2007. http://dx.doi.org/10.1074/jbc.M700146200
- J. Hahn, and D.J. Thiele, "Activation of the Saccharomyces cerevisiae Heat Shock Transcription Factor Under Glucose Starvation Conditions by Snf1 Protein Kinase", Journal of Biological Chemistry, vol. 279, pp. 5169-5176, 2004. http://dx.doi.org/10.1074/jbc.M311005200
- M. Platara, A. Ruiz, R. Serrano, A. Palomino, F. Moreno, and J. Ariño, "The Transcriptional Response of the Yeast Na+-ATPase ENA1 Gene to Alkaline Stress Involves Three Main Signaling Pathways", Journal of Biological Chemistry, vol. 281, pp. 36632-36642, 2006. http://dx.doi.org/10.1074/jbc.M606483200
- K. Mori, "Signalling Pathways in the Unfolded Protein Response: Development from Yeast to Mammals", Journal of Biochemistry, vol. 146, pp. 743-750, 2009. http://dx.doi.org/10.1093/jb/mvp166
- P. Walter, and D. Ron, "The Unfolded Protein Response: From Stress Pathway to Homeostatic Regulation", Science, vol. 334, pp. 1081-1086, 2011. http://dx.doi.org/10.1126/science.1209038
- J. Ferrer-Dalmau, F. Randez-Gil, M. Marquina, J. Prieto, and A. Casamayor, "Protein kinase Snf1 is involved in the proper regulation of the unfolded protein response in Saccharomyces cerevisiae", Biochemical Journal, vol. 468, pp. 33-47, 2015. http://dx.doi.org/10.1042/BJ20140734
- T. Mizuno, Y. Masuda, and K. Irie, "The Saccharomyces cerevisiae AMPK, Snf1, Negatively Regulates the Hog1 MAPK Pathway in ER Stress Response", PLOS Genetics, vol. 11, pp. e1005491, 2015. http://dx.doi.org/10.1371/journal.pgen.1005491
- S.H. Back, M. Schröder, K. Lee, K. Zhang, and R.J. Kaufman, "ER stress signaling by regulated splicing: IRE1/HAC1/XBP1", Methods, vol. 35, pp. 395-416, 2005. http://dx.doi.org/10.1016/J.YMETH.2005.03.001
- K. Simpson-Lavy, A. Bronstein, M. Kupiec, and M. Johnston, "Cross-Talk between Carbon Metabolism and the DNA Damage Response in S. cerevisiae", Cell Reports, vol. 12, pp. 1865-1875, 2015. http://dx.doi.org/10.1016/j.celrep.2015.08.025
- A. Salminen, K. Kaarniranta, and A. Kauppinen, "Age-related changes in AMPK activation: Role for AMPK phosphatases and inhibitory phosphorylation by upstream signaling pathways", Ageing Research Reviews, vol. 28, pp. 15-26, 2016. http://dx.doi.org/10.1016/j.arr.2016.04.003
- R. Friis, J. Glaves, T. Huan, L. Li, B. Sykes, and M. Schultz, "Rewiring AMPK and Mitochondrial Retrograde Signaling for Metabolic Control of Aging and Histone Acetylation in Respiratory-Defective Cells", Cell Reports, vol. 7, pp. 565-574, 2014. http://dx.doi.org/10.1016/j.celrep.2014.03.029
- D.R. Lorenz, C.R. Cantor, and J.J. Collins, "A network biology approach to aging in yeast", Proceedings of the National Academy of Sciences, vol. 106, pp. 1145-1150, 2009. http://dx.doi.org/10.1073/pnas.0812551106
- J. Apfeld, G. O'Connor, T. McDonagh, P.S. DiStefano, and R. Curtis, "The AMP-activated protein kinase AAK-2 links energy levels and insulin-like signals to lifespan in C. elegans", Genes & Development, vol. 18, pp. 3004-3009, 2004. http://dx.doi.org/10.1101/gad.1255404
-
S. Yang, L. Long, D. Li, J. Zhang, S. Jin, F. Wang, and J. Chen, "β‐Guanidinopropionic acid extends the lifespan of
D rosophila melanogaster via anAMP ‐activated protein kinase‐dependent increase in autophagy", Aging Cell, vol. 14, pp. 1024-1033, 2015. http://dx.doi.org/10.1111/acel.12371 - R.M. Reznick, and G.I. Shulman, "The role of AMP‐activated protein kinase in mitochondrial biogenesis", The Journal of Physiology, vol. 574, pp. 33-39, 2006. http://dx.doi.org/10.1113/jphysiol.2006.109512
- J. Kim, M. Kundu, B. Viollet, and K. Guan, "AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1", Nature Cell Biology, vol. 13, pp. 132-141, 2011. http://dx.doi.org/10.1038/ncb2152
- H. Kim, S.Y. Moon, J. Kim, C.H. Baek, M. Kim, J.Y. Min, and S.K. Lee, "Activation of AMP-activated protein kinase inhibits ER stress and renal fibrosis", American Journal of Physiology-Renal Physiology, vol. 308, pp. F226-F236, 2015. http://dx.doi.org/10.1152/ajprenal.00495.2014
- T. Sanli, G.R. Steinberg, G. Singh, and T. Tsakiridis, "AMP-activated protein kinase (AMPK) beyond metabolism", Cancer Biology & Therapy, vol. 15, pp. 156-169, 2013. http://dx.doi.org/10.4161/cbt.26726
- R.M. Reznick, H. Zong, J. Li, K. Morino, I.K. Moore, H.J. Yu, Z. Liu, J. Dong, K.J. Mustard, S.A. Hawley, D. Befroy, M. Pypaert, D.G. Hardie, L.H. Young, and G.I. Shulman, "Aging-Associated Reductions in AMP-Activated Protein Kinase Activity and Mitochondrial Biogenesis", Cell Metabolism, vol. 5, pp. 151-156, 2007. http://dx.doi.org/10.1016/j.cmet.2007.01.008
- M.B. Wierman, N. Maqani, E. Strickler, M. Li, and J.S. Smith, "Caloric Restriction Extends Yeast Chronological Life Span by Optimizing the Snf1 (AMPK) Signaling Pathway", Molecular and Cellular Biology, vol. 37, 2017. http://dx.doi.org/10.1128/MCB.00562-16
- N. Maqani, R.D. Fine, M. Shahid, M. Li, E. Enriquez-Hesles, and J.S. Smith, "Spontaneous mutations in CYC8 and MIG1 suppress the short chronological lifespan of budding yeast lacking SNF1/AMPK", Microbial Cell, vol. 5, pp. 233-248, 2018. http://dx.doi.org/10.15698/mic2018.05.630
-
T. Ogawa, R. Tsubakiyama, M. Kanai, T. Koyama, T. Fujii, H. Iefuji, T. Soga, K. Kume, T. Miyakawa, D. Hirata, and M. Mizunuma, "Stimulating
S
-adenosyl-
l -methionine synthesis extends lifespan via activation of AMPK", Proceedings of the National Academy of Sciences, vol. 113, pp. 11913-11918, 2016. http://dx.doi.org/10.1073/pnas.1604047113 - C. López-Otín, L. Galluzzi, J.M. Freije, F. Madeo, and G. Kroemer, "Metabolic Control of Longevity", Cell, vol. 166, pp. 802-821, 2016. http://dx.doi.org/10.1016/j.cell.2016.07.031
- V.D. Longo, and P. Fabrizio, "Chronological Aging in Saccharomyces cerevisiae", Aging Research in Yeast, pp. 101-121, 2011. http://dx.doi.org/10.1007/978-94-007-2561-4_5
- P. Laun, M. Rinnerthaler, E. Bogengruber, G. Heeren, and M. Breitenbach, "Yeast as a model for chronological and reproductive aging – A comparison", Experimental Gerontology, vol. 41, pp. 1208-1212, 2006. http://dx.doi.org/10.1016/j.exger.2006.11.001
- M. Becuwe, N. Vieira, D. Lara, J. Gomes-Rezende, C. Soares-Cunha, M. Casal, R. Haguenauer-Tsapis, O. Vincent, S. Paiva, and S. Léon, "A molecular switch on an arrestin-like protein relays glucose signaling to transporter endocytosis", Journal of Cell Biology, vol. 196, pp. 247-259, 2012. http://dx.doi.org/10.1083/jcb.201109113
- V. Llopis-Torregrosa, A. Ferri-Blázquez, A. Adam-Artigues, E. Deffontaines, G.H. van Heusden, and L. Yenush, "Regulation of the Yeast Hxt6 Hexose Transporter by the Rod1 α-Arrestin, the Snf1 Protein Kinase, and the Bmh2 14-3-3 Protein", Journal of Biological Chemistry, vol. 291, pp. 14973-14985, 2016. http://dx.doi.org/10.1074/jbc.M116.733923
- A.F. O'Donnell, R.R. McCartney, D.G. Chandrashekarappa, B.B. Zhang, J. Thorner, and M.C. Schmidt, "2-Deoxyglucose Impairs Saccharomyces cerevisiae Growth by Stimulating Snf1-Regulated and α-Arrestin-Mediated Trafficking of Hexose Transporters 1 and 3", Molecular and Cellular Biology, vol. 35, pp. 939-955, 2015. http://dx.doi.org/10.1128/MCB.01183-14
- J. Hsu, K. Chen, and F.S. Lee, "Snf1/AMP-activated protein kinase activates Arf3p to promote invasive yeast growth via a non-canonical GEF domain", Nature Communications, vol. 6, 2015. http://dx.doi.org/10.1038/ncomms8840
- S. Kuchin, V.K. Vyas, and M. Carlson, "Snf1 Protein Kinase and the Repressors Nrg1 and Nrg2 Regulate FLO11, Haploid Invasive Growth, and Diploid Pseudohyphal Differentiation", Molecular and Cellular Biology, vol. 22, pp. 3994-4000, 2002. http://dx.doi.org/10.1128/MCB.22.12.3994-4000.2002
- V.K. Vyas, S. Kuchin, C.D. Berkey, and M. Carlson, "Snf1 Kinases with Different β-Subunit Isoforms Play Distinct Roles in Regulating Haploid Invasive Growth", Molecular and Cellular Biology, vol. 23, pp. 1341-1348, 2003. http://dx.doi.org/10.1128/MCB.23.4.1341-1348.2003
- C.L. Denis, "Identification of new genes involved in the regulation of yeast alcohol dehydrogenase II.", Genetics, 1984. http://www.ncbi.nlm.nih.gov/pubmed/6392016
-
F. Galello, C. Pautasso, S. Reca, L. Cañonero, P. Portela, S. Moreno, and S. Rossi, "Transcriptional regulation of the protein kinase a subunits in
Saccharomyces cerevisiae during fermentative growth", Yeast, vol. 34, pp. 495-508, 2017. http://dx.doi.org/10.1002/yea.3252 - S. Busnelli, F. Tripodi, R. Nicastro, C. Cirulli, G. Tedeschi, R. Pagliarin, L. Alberghina, and P. Coccetti, "Snf1/AMPK promotes SBF and MBF-dependent transcription in budding yeast", Biochimica et Biophysica Acta (BBA) - Molecular Cell Research, vol. 1833, pp. 3254-3264, 2013. http://dx.doi.org/10.1016/j.bbamcr.2013.09.014
- P.T. Spellman, G. Sherlock, M.Q. Zhang, V.R. Iyer, K. Anders, M.B. Eisen, P.O. Brown, D. Botstein, and B. Futcher, "Comprehensive Identification of Cell Cycle–regulated Genes of the YeastSaccharomyces cerevisiaeby Microarray Hybridization", Molecular Biology of the Cell, vol. 9, pp. 3273-3297, 1998. http://dx.doi.org/10.1091/mbc.9.12.3273
- R.R. McCartney, D.G. Chandrashekarappa, B.B. Zhang, and M.C. Schmidt, "Genetic Analysis of Resistance and Sensitivity to 2-Deoxyglucose in Saccharomyces cerevisiae", Genetics, vol. 198, pp. 635-646, 2014. http://dx.doi.org/10.1534/genetics.114.169060
- F. Tripodi, R. Fraschini, M. Zocchi, V. Reghellin, and P. Coccetti, "Snf1/AMPK is involved in the mitotic spindle alignment in Saccharomyces cerevisiae", Scientific Reports, vol. 8, 2018. http://dx.doi.org/10.1038/s41598-018-24252-y
- J.K. Moore, and J.A. Cooper, "Coordinating mitosis with cell polarity: Molecular motors at the cell cortex", Seminars in Cell & Developmental Biology, vol. 21, pp. 283-289, 2010. http://dx.doi.org/10.1016/j.semcdb.2010.01.020
- R. Nicastro, F. Tripodi, C. Guzzi, V. Reghellin, S. Khoomrung, C. Capusoni, C. Compagno, C. Airoldi, J. Nielsen, L. Alberghina, and P. Coccetti, "Enhanced amino acid utilization sustains growth of cells lacking Snf1/AMPK", Biochimica et Biophysica Acta (BBA) - Molecular Cell Research, vol. 1853, pp. 1615-1625, 2015. http://dx.doi.org/10.1016/j.bbamcr.2015.03.014
- S. Jeon, "Regulation and function of AMPK in physiology and diseases", Experimental & Molecular Medicine, vol. 48, pp. e245-e245, 2016. http://dx.doi.org/10.1038/emm.2016.81
-
D. Grahame Hardie, "
AMP ‐activated protein kinase: a key regulator of energy balance with many roles in human disease", Journal of Internal Medicine, vol. 276, pp. 543-559, 2014. http://dx.doi.org/10.1111/joim.12268 - B. Faubert, G. Boily, S. Izreig, T. Griss, B. Samborska, Z. Dong, F. Dupuy, C. Chambers, B. Fuerth, B. Viollet, O. Mamer, D. Avizonis, R. DeBerardinis, P. Siegel, and R. Jones, "AMPK Is a Negative Regulator of the Warburg Effect and Suppresses Tumor Growth In Vivo", Cell Metabolism, vol. 17, pp. 113-124, 2013. http://dx.doi.org/10.1016/j.cmet.2012.12.001
- B. Faubert, E.E. Vincent, M.C. Poffenberger, and R.G. Jones, "The AMP-activated protein kinase (AMPK) and cancer: Many faces of a metabolic regulator", Cancer Letters, vol. 356, pp. 165-170, 2015. http://dx.doi.org/10.1016/j.canlet.2014.01.018
- S. Jeon, N.S. Chandel, and N. Hay, "AMPK regulates NADPH homeostasis to promote tumour cell survival during energy stress", Nature, vol. 485, pp. 661-665, 2012. http://dx.doi.org/10.1038/nature11066
ACKNOWLEDGMENTS
Work in the authors' laboratory was supported by SysBioNet project, a MIUR initiative from the Italian Roadmap of European Strategy Forum on Research Infrastructures (ESFRI). F.T. has been supported by fellowships from MIUR, R.N. was funded by a fellowship from SysBioNet.
COPYRIGHT
© 2018

Conventional and emerging roles of the energy sensor Snf1/AMPK in Saccharomyces cerevisiae by Coccetti et al. is licensed under a Creative Commons Attribution 4.0 International License.