Back to article: Fat storage-inducing transmembrane (FIT or FITM) proteins are related to lipid phosphatase/phosphotransferase enzymes
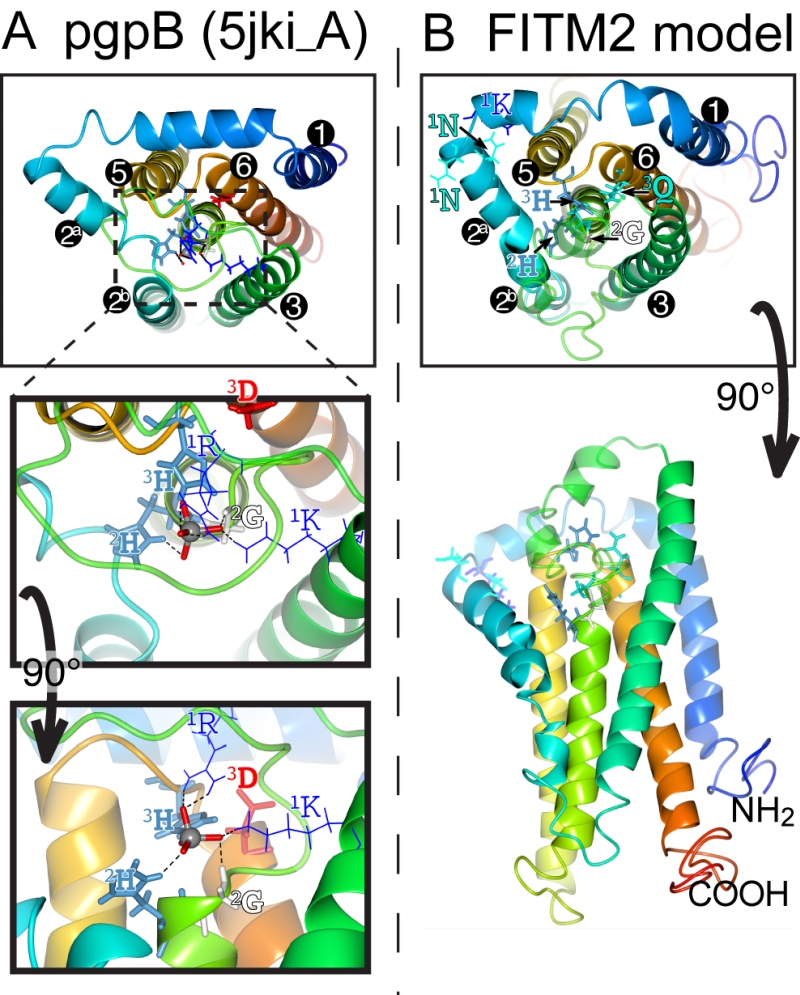
FIGURE 8: Structural model of FITM2. (A) Lumenal aspect of bacterial PgpB enzyme (accession 5jki, bottom panel), which is the highest scoring threading template for FITM2 identified by I-TASSER, numbering its 6 TMDs, except the central TMD 4, and with TMD2 split in 2 parts. Zoom in (middle panel) and rotation to side aspect (top panel) show the highlighted elements of the active site, including the tungsten ion (gray sphere) crystallised in place of the phosphate group in 5jki, and the side chains of co-ordinating conserved residues (coloured by residue type) in C1 (1K, 1R = thin bonds), C2 and C3 (2G, 2H, 3H, 3D = fat bonds) [74]. (B) Superimposed view of the modelled structure of human FITM2 with side-chains of the conserved residues in C2 and C3 as in A (bottom panel = lumenal aspect, top panel = side aspect), plus the hydrophilic residues (KxNxxN) conserved in loop 1/2 (thin bonds). Models were obtained from the I-TASSER server for 3D structure prediction with standard settings [75]. Several models with seven TMDs were rejected on the basis of topology studies of Yft2p and Scs3p showing six TMDs [13]. In all panels, colouring of the chain graduates from blue (N-terminus) to red (C-terminus).
13. Gross DA, Snapp EL, Silver DL (2010). Structural insights into triglyceride storage mediated by fat storage-inducing transmembrane (FIT) protein 2. PLoS One 5(5): e10796. http://dx.doi.org/10.1371/journal.pone.0010796
58. Ghachi ME, Howe N, Auger R, Lambion A, Guiseppi A, Delbrassine F, Manat G, Roure S, Peslier S, Sauvage E, Vogeley L, Rengifo-Gonzalez JC, Charlier P, Mengin-Lecreulx D, Foglino M, Touze T, Caffrey M, Kerff F (2017). Crystal structure and biochemical characterization of the transmembrane PAP2 type phosphatidylglycerol phosphate phosphatase from Bacillus subtilis. Cell Mol Life Sci 74(12): 2319-2332. 10.1007/s00018-017-2464-6
59. Roy A, Kucukural A, Zhang Y (2010). I-TASSER: a unified platform for automated protein structure and function prediction. Nature protocols 5(4): 725-738. http://dx.doi.org/10.1038/nprot.2010.5