Microreviews:
Microbial Cell, Vol. 1, No. 5, pp. 158 - 159; doi: 10.15698/mic2014.05.144
Prokaryotic Argonautes – variations on the RNA interference theme
1 Laboratory of Microbiology, Wageningen University, Dreijenplein 10, 6703 HB Wageningen, Netherlands.
2 Sir William Dunn School of Pathology, University of Oxford, South Parks Road, Oxford OX1 3RE, UK.
Keywords: Argonaute, RNAi, Bacteria, Archaea.
Received originally: 26/03/2014 Accepted: 28/03/2014
Published: 15/04/2014
Correspondence:
John van der Oost, Dreijenplein 10; 6703 HB Wageningen, Netherlands john.vanderoost@wur.nl
Conflict of interest statement: The authors declare no competing financial interests.
Please cite this article as: John van der Oost, Daan C. Swarts, Matthijs M. Jore (2014). Prokaryotic Argonautes – variations on the RNA interference theme. Microbial Cell 1(5): 158-159.
The discovery of RNA interference (RNAi) has been a major scientific breakthrough. This RNA-guided RNA interference system plays a crucial role in a wide range of regulatory and defense mechanisms in eukaryotes. The key enzyme of the RNAi system is Argonaute (Ago), an endo-ribonuclease that uses a small RNA guide molecule to specifically target a complementary RNA transcript. Two functional classes of eukaryotic Ago have been described: catalytically active Ago that cleaves RNA targets complementary to its guide, and inactive Ago that uses its guide to bind target RNA to down-regulate translation efficiency. A recent comparative genomics study has revealed that Argonaute-like proteins are also encoded by prokaryotic genomes. Interestingly, there is a lot of variation among these prokaryotic Argonaute (pAgo) proteins with respect to domain architecture: some resemble the eukaryotic Ago (long pAgo) containing a complete or disrupted catalytic site, while others are truncated versions (short pAgo) that generally contain an incomplete catalytic site. Prokaryotic Agos with an incomplete catalytic site often co-occur with (predicted) nucleases. Based on this diversity, and on the fact that homologs of other RNAi-related protein components (such as Dicer nucleases) have never been identified in prokaryotes, it has been predicted that variations on the eukaryotic RNAi theme may occur in prokaryotes.
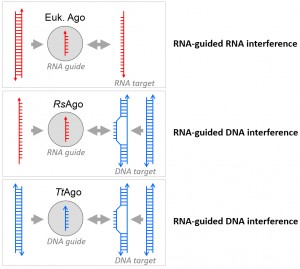
Recent studies by us and the working group of Alexei Aravin have described molecular analyses of two distinct bacterial Argonautes, TtAgo from Thermus thermophilus and RsAgo from Rhodobacter sphaeroides. Like eukaryotic Argonautes, both TtAgo and RsAgo are long pAgos that are co-purified with oligonucleotide guides (13-25 nt and 15-19 nt, respectively), the majority of which are complementary to plasmids. Together with the observation that TtAgo (and most likely also RsAgo) target DNA, this has led to the conclusion that both pAgos play a role in host defense (Figure 1). However, apart from these similarities there are important functional differences between the two bacterial Argonaute proteins. The guides acquired by RsAgo are mRNA-derived RNA oligonucleotides that target the template strand of plasmid genes. As RsAgo lacks a catalytic site, it most likely requires a partner nuclease for target cleavage. In contrast, TtAgo acquires DNA guides that allow targeting of AT-rich sequences of double-stranded plasmid DNA. Whereas the functional nuclease site of TtAgo catalyzes nicking of a single targeted DNA strand, two TtAgos loaded with overlapping complementary guides can generate double-stranded DNA breaks.
These studies have revealed interesting variations on the eukaryotic RNAi theme. Several basic features of these two variant pAgos remain elusive, concerning mechanistic details of both guide acquisition and target interference. Moreover, apart from the two characterized prokaryotic Argonautes, many more pAgo variants exist, that may differ in functionality with respect to (i) guide preference (RNA/DNA), (ii) target specificity (RNA/DNA), and (iii) catalytic mechanism (nuclease activity). Apart from providing insights in the evolution of the prokaryotic Ago variants and their eukaryotic counterparts, future research will aim at revealing the molecular basis for the distinct functionality of these different pAgo variants. Moreover, gained insights will result in an interesting set of novel nucleases that may allow for dedicated genetic engineering.
COPYRIGHT
© 2014

Prokaryotic Argonautes – variations on the RNA interference theme by John van der Oost et al. is licensed under a Creative Commons Attribution 4.0 International License.