Back to article: Sulfur transfer and activation by ubiquitin-like modifier system Uba4•Urm1 link protein urmylation and tRNA thiolation in yeast
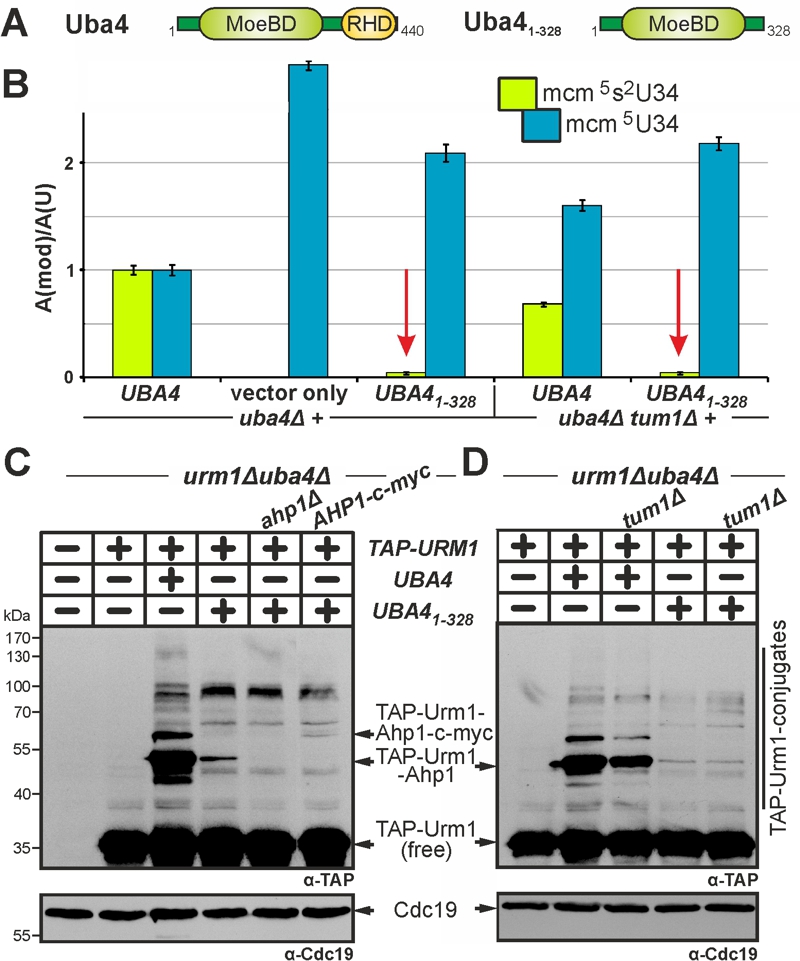
FIGURE 5: Low residual tRNA thiolation and urmylation without the RHD on Uba4 is independent of sulfur transfer by Tum1.
(A) Scheme depicting Uba41‑328, which lacks the RHD region.
(B) LC-MS/MS-based analysis (see Fig. 3) reveals Uba41‑328 is severely compromised in tRNA thiolation (red arrows) with the remaining residual (~4%) levels of mcm5s2-modified U34 insensitive to TUM1 gene function.
(C, D) Urmylation of proteins including Ahp1 is strongly suppressed in the absence of the RHD. Urmylation assays on protein extracts from the indicated genetic backgrounds involved anti-TAP-based EMSA (see Fig. 2) and immune blots to detect non-conjugated (free) TAP-Urm1 and TAP-Urm1 conjugates (C, D) including urmylated forms of wild-type (C, D) and c-myc-tagged Ahp1 (D). Protein loading (C, D) was controlled with anti-Cdc19 Western blots. Note that as is the case with very weak Tum1 insensitive tRNA thiolation (B), low residual levels of urmylation with Uba41‑328 are also unaltered in the tum1Δ mutant (D).