Back to article: Forced association of SARS-CoV-2 proteins with the yeast proteome perturb vesicle trafficking
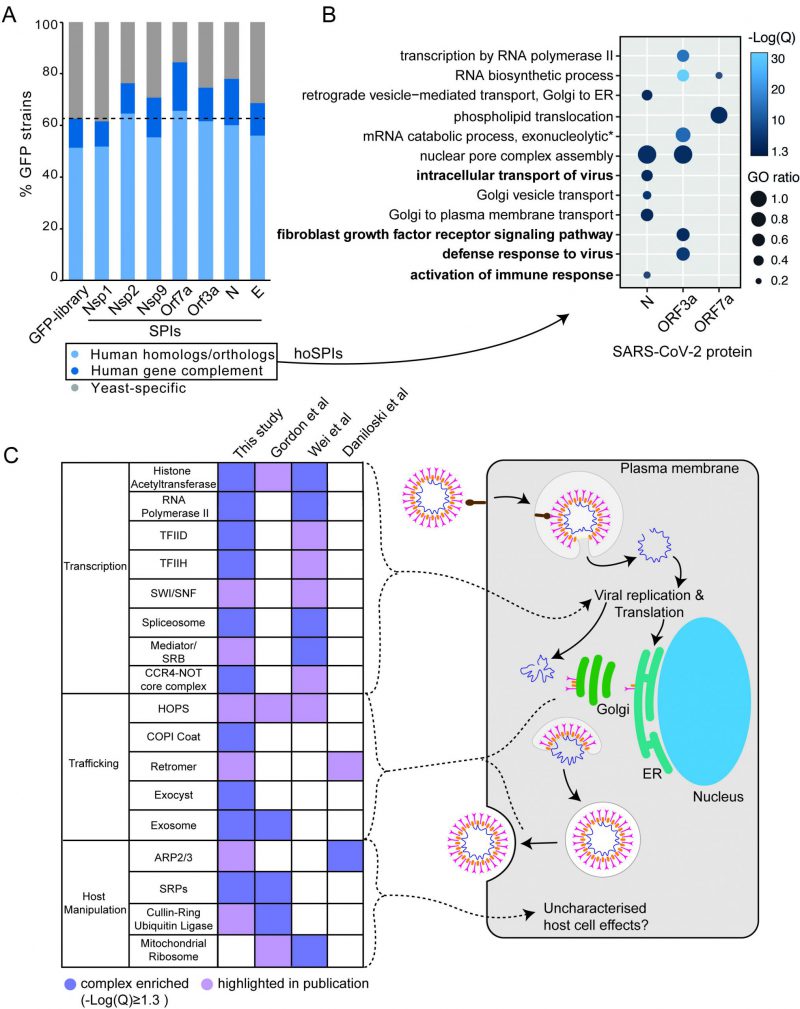
FIGURE 4: SARS-CoV-2 SPI screens in the context of human genetics. (A) The SARS-CoV-2 yeast SPIs are collectively enriched for those that have human homologues or orthologues compared to the GFP strains tested (Fishers exact test, p=7.8×10-8). Human Gene Complement indicates those human genes that have been demonstrated to complement the loss of the yeast homolog/ortholog. (B) Enrichment analysis of GO biological process terms of human orthologues of the SARS-CoV-2 SPIs (hoSPIs). Bold terms indicate human specific GO terms, that are not present in yeast. The -Log(Q) value is illustrated by blue shading, all -Log(Q) values are greater than or equal to 1.3, which is equivalent to q-values < 0.05. GO ratio (size of the circle) indicates the proportion of genes in a GO term were found for each enrichment. (C) A comparison of the complexes that contain hoSPIs with those identified from three SARS-CoV-2 screens performed in mammalian cells. Dark purple shading indicates enrichment, light purple shading highlights complexes which are mentioned by the authors. These complexes correspond to the various steps in the viral lifecycle (right).