Research Articles:
Microbial Cell, Vol. 11, No. 1, pp. 339 - 352; doi: 10.15698/mic2024.10.837
RidA proteins contribute to fitness of S. enterica and E. coli by reducing 2AA stress and moderating flux to isoleucine biosynthesis
Department of Microbiology, University of Georgia, Athens, GA 30602-2605.
Keywords: RidA, 2-aminoacrylate, competitive fitness, metabolic stress.
Received originally: 31/05/2024 Received in revised form: 22/08/2024
Accepted: 29/08/2024
Published: 04/10/2024
Correspondence:
Dr. Diana M Downs, Department of Microbiology, University of Georgia, Athens, GA, USA; dmdowns@uga.edu
Conflict of interest statement: The authors declare no conflicts of interest.
Please cite this article as: Ronnie L. Fulton, Bryce R. Sawyer, Diana M Downs (2024). RidA proteins contribute to fitness of S. enterica and E. coli by reducing 2AA stress and moderating flux to isoleucine biosynthesis. Microbial Cell 11: 339-352. doi: 10.15698/mic2024.10.837
Abstract
Defining the physiological role of a gene product relies on interpreting phenotypes caused by the lack, or alteration, of the respective gene product. Mutations in critical genes often lead to easily recognized phenotypes that can include changes in cellular growth, metabolism, structure etc. However, mutations in many important genes may fail to generate an obvious defect unless additional perturbations are caused by medium or genetic background. The latter scenario is exemplified by RidA proteins. In vitro RidA proteins deaminate numerous imine/enamines, including those generated by serine/threonine dehydratase IlvA (EC:4.3.1.19) from serine or threonine – 2-aminoacrylate (2AA) and 2- aminocrotonate (2AC), respectively. Despite this demonstrable biochemical activity, a lack of RidA has little to no effect on growth of E. coli or S. enterica without the application of additional metabolic perturbation. A cellular role of RidA is to prevent accumulation of 2AA which, if allowed to persist, can irreversibly damage pyridoxal 5’-phosphate (PLP)-dependent enzymes, causing global metabolic stress. Because the phenotypes caused by a lack of RidA are dependent on the unique structure of each metabolic network, the link between RidA function and 2AA stress is difficult to demonstrate in some organisms. The current study used coculture experiments to exacerbate differences in growth caused by the lack of RidA in S. enterica and E. coli. Results described here solidify the established role of RidA in removing 2AA, while also presenting evidence for a role of RidA in enhancing flux towards isoleucine biosynthesis in E. coli. Overall, these data emphasize that metabolic networks can generate distinct responses to perturbation, even when the individual components are conserved.
INTRODUCTION
The robust metabolic network of a microbial cell requires the integration of individual components (i.e., enzymes and pathways) and multi-layered regulation of the resulting connections. In a general sense, the metabolic network is an amalgamation of interconnected biochemical pathways which are themselves composed of a diverse repertoire of enzymes. Cells modulate flux through these pathways by altering the expression or activity of appropriate enzymes which, in turn, dictate the rate and direction that relevant metabolites flow through the network. Precise control over cellular metabolism is allowed by the structural complexity of the system, emphasizing the notion that the individual parts have evolved to function within the context of the entire network. The precision allows microbial cells to survive in dynamic environments by restoring stability after a perturbation has been applied to the network.
Numerous components of metabolic networks in model bacteria are well characterized, although many enzymes and pathways remain poorly or only partially described. Historically, a first step in identifying the role of a gene is an analysis of phenotype(s) caused by the absence of the respective gene product. A detectable mutant phenotype demonstrates a physiological need for the gene and provides a tool to unravel the molecular function of the gene product. This approach loses its utility when a mutant has no detectable phenotype to guide functional studies. As a consequence, numerous genes remain uncharacterized, even in the best studied model systems. For example, the genome of Escherichia coli contains the coding capacity for over 4500 genes, an estimated 1200 of which have no associated phenotype or known function 1. The characterization of these genes requires innovative experimental design and/or integration of data from genetic, biochemical and global analyses.
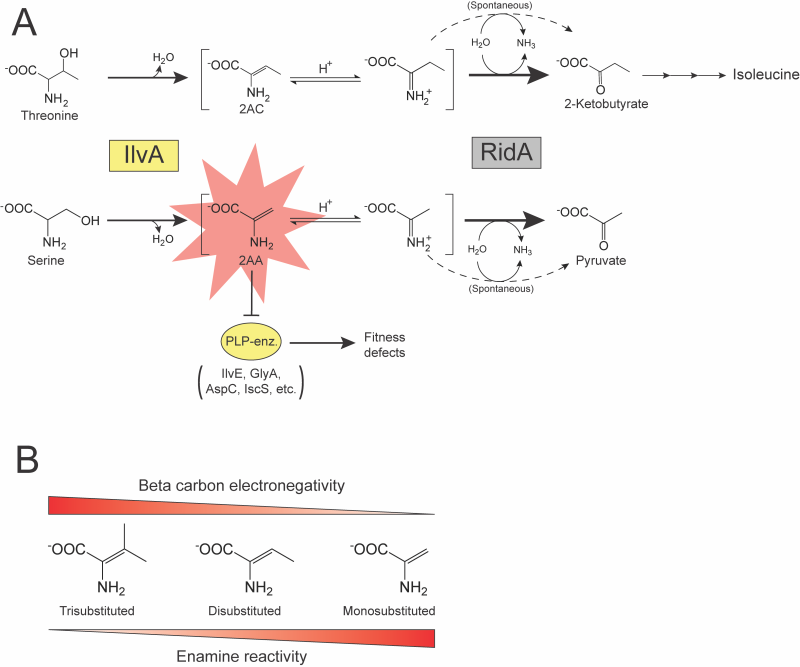
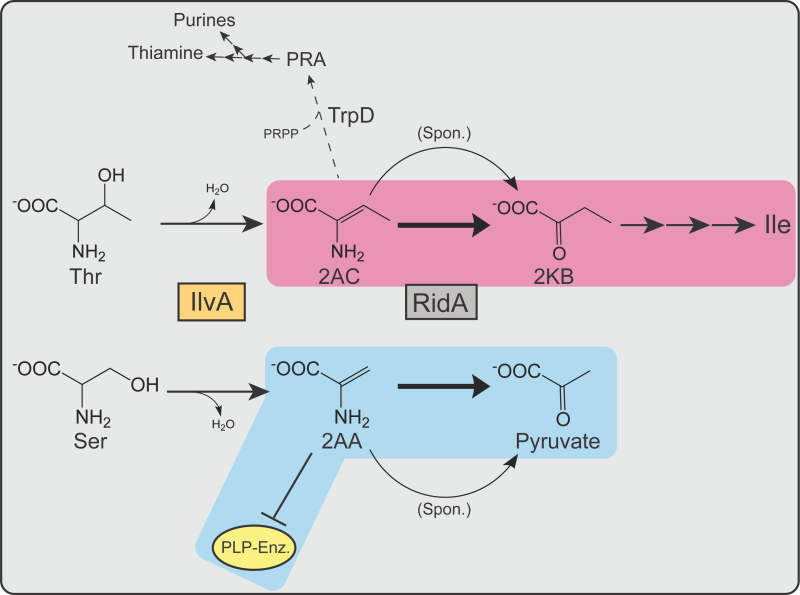
Studies of the Rid family of proteins exemplify the difficulty in characterizing genes whose absence fails to cause a detectable phenotype in the laboratory. The Rid superfamily is divided into eight subfamilies 2, of which the RidA subfamily is the best characterized. In the past two decades, numerous experiments have probed the function of RidA across multiple model bacterial species. These studies identified RidA as an enamine/imine deaminase with at least two substrates of physiological significance, 2-aminoacrylate (2AA) and 2-aminocrotonate (2AC). The phenotypes of a ridA mutant that led to its functional characterization required further metabolic perturbations 3, 4, 5, or additional mutations 6, 7, 8. The serine/threonine dehydratase IlvA (EC:4.3.1.19) generates 2AA and 2AC via the dehydration of serine or threonine, respectively 3, 9. Past studies, primarily in Salmonella enterica, demonstrated that 2AA and 2AC accumulate in the absence of RidA. 2AA has been of particular interest due to its increased reactivity and toxicity compared to 2AC. 2AA has an unsubstituted pi bond between the α- and β-carbons, which is less stable than the disubstituted pi bond of 2AC (Figure 1B). This feature makes the β-carbon of 2AA less electronegative, which will favor tautomerization and allow subsequent nucleophilic attack by this molecule 10. The reactivity of 2AA facilitates attack on the pyridoxal 5’-phosphate (PLP) in the active site of some enzymes. Stress caused by 2AA accumulation results from decreased activity of several PLP-dependent enzymes that can cause growth defects in a ridA mutant (Figure 1A).
–
Over time, the RidA/2AA paradigm was described, target enzymes and metabolic consequences were identified, and suppressor mutations were isolated that minimized the impact of 2AA 11, 12, 13, 14, 15, 16, 17, 18. Features of this paradigm are present in several organisms, which led to the assumption that 2AA deaminase activity of RidA was the driver of its conservation and broad phylogenetic distribution 19, 20.
Recent observations have suggested that the deamination of 2AA may not be the sole cellular function of RidA. For instance, in some organisms (Bacillus subtilis, E. coli) no evidence of naturally generated 2AA stress has been identified 21 (Buckner, unpublished). Further, RidA proteins deaminate diverse substrates in vitro, suggesting there may be additional physiologically relevant molecules acted on by RidA 20. The latter possibility was prompted by the role of non-RidA members of the Rid family in deaminating imines that are not known to damage cellular components in a similar fashion as 2AA 22, 23, 24. All reactions currently attributed to Rid proteins can also proceed non-enzymatically, if catalyzed by free water. However, in all described cases, the presence of a Rid protein increases the rate of product formation over that of the reaction catalyzed by water 2, 3, 19, 22, 23, 24, 25, 26, 27, 28. In the case of RidA, the enhanced removal of 2AA ameliorates metabolic stress. An emerging pattern suggests that members of the Rid superfamily function to deaminate metabolic intermediates and increase flux through a particular metabolic pathway 2, 19, 22, 23, 24, 26. For example, the Rid protein RutC is encoded in the pyrimidine utilization operon of E. coli and catalyzes the deamination of 3-aminoacrylate to malonate semialdehyde in a reaction previously designated as spontaneous 24, 29. Additionally, members of the Rid2 subfamily in Pseudomonas species enhance flux through amino acid catabolic pathways, providing a competitive fitness advantage over strains lacking the Rid protein 22, 23.
The current study addressed the cellular role of RidA in S. enterica and E. coli in the context of a possible role in removing 2AC. 2AC is a physiologically relevant substrate of RidA at least in S. enterica and is not known to have toxic consequences 3, 8. 2AC is an obligatory intermediate in the biosynthesis of isoleucine generated from threonine by IlvA. Results obtained from coculture experiments described herein confirm a role of RidA in preventing 2AA stress and suggest that RidA proteins may also have an organism-specific role in modulating biosynthetic flux to isoleucine.
RESULTS
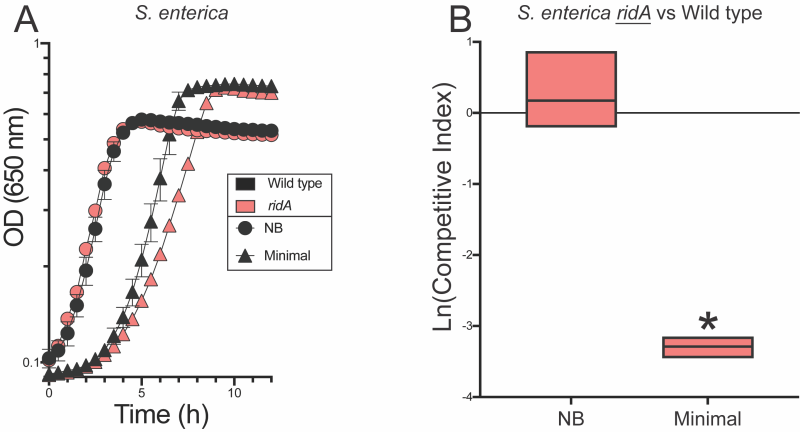
S. enterica ridA mutants have a defect in competitive fitness
In minimal glucose medium, a ridA mutant has a slight, often immeasurable, growth defect in monoculture (Figure 2, Figure 3). Despite the lack of a significant growth phenotype, activity of branched chain aminotransferase (IlvE, EC:2.6.1.42) is decreased by ~40% in a ridA mutant under these conditions. This demonstrable biochemical defect indicates 2AA has accumulated 30, 31, 32. Serine can be added to the medium to increase 2AA stress. The presence of serine prevents growth of a ridA mutant in S. enterica by partially inactivating serine hydroxymethyltransferase (GlyA, EC:2.1.2.1) and, to a lesser extent, cysteine desulfurase (IscS, EC:2.8.1.7) 5, 13, 18. In competition experiments, S. enterica ridA mutants and wild type cells were co-inoculated into nutrient medium and minimal glucose medium. The competitive index (CI) for strains was calculated as the ratio of strains after growth in coculture normalized to the ratio present in the inoculum. The CI is a measure of competitive fitness that has typically been used to study the requirement for gene products when cells are grown in competition for a single limiting resource 22, 23. In contrast, experiments herein were designed to define competition under conditions where no single resource was limiting. In other words, CI values presented here define differences in general fitness (i.e., growth rate) of a strain that may not be apparent in monoculture experiments. No difference in fitness was detected between wild type and a strain lacking RidA grown in nutrient medium. In contrast, the ridA mutant had a significant fitness defect when the two strains were cocultured in minimal glucose medium. In minimal medium, the ridA mutant had a CI of 0.037 (ln(CI) = -3.3), which reflects a ~27-fold decrease in fitness compared to wild type (Figure 2). The significant fitness difference between wild type S. enterica and the ridA mutant amplified a growth difference that was minimal, or undetectable, in monoculture and underlined the sensitivity of this approach. Results from this competition assay aligned with the 2AA-dependent decrease in IlvE activity of a ridA mutant grown in minimal medium, validating that 2AA stress is present in S. enterica during growth in minimal medium 30, 31.
–
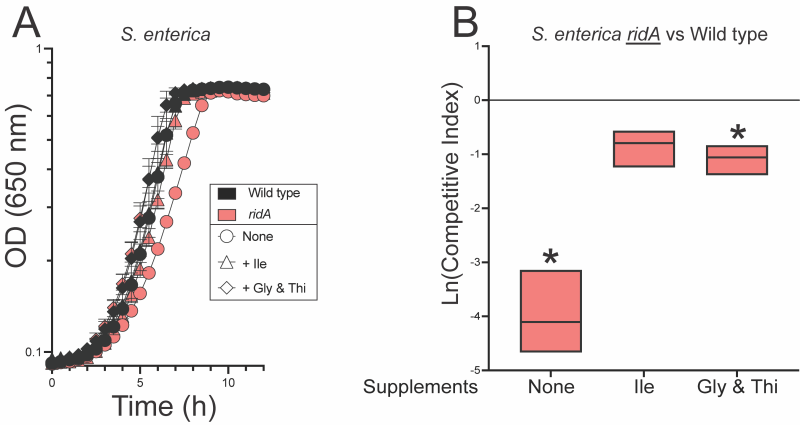
Isoleucine restores fitness of a S. enterica ridA mutant in coculture
Isoleucine in the growth medium restored full competitive fitness to the ridA mutant (Figure 3). This result was expected, as isoleucine is known to allosterically inhibit serine/threonine dehydratase IlvA and prevent the formation of 2AA 30, 31. These data supported the idea that the fitness defect of a ridA mutant was due to accumulated 2AA.
–
A caveat to this simple interpretation is that RidA also deaminates the threonine-derived enamine product of IlvA, 2AC. 2AC is an obligatory intermediate in the biosynthesis of isoleucine that is eliminated when isoleucine inhibits IlvA. Thus, it was possible that RidA improved fitness of S. enterica by increasing biosynthetic flux to isoleucine. Distinguishing these two possibilities was complicated by the fact that isoleucine would eliminate the need for RidA in both scenarios. Coculture experiments to separate the possible contributions of RidA were based on the response of S. enterica to higher levels of 2AA stress generated when exogenous serine was added. The assumption was that the effect of 2AA stress generated by endogenous serine would be similar in consequence, if not magnitude, to that generated by addition of exogenous serine. Specifically, S. enterica ridA mutants grow well in the presence of 2AA stress if glycine and thiamine are provided. Together, these nutrients bypass the two key enzymes damaged by 2AA, GlyA and IscS and allow growth in the presence of 2AA stress 5, 13. Notably, the presence of glycine and thiamine in the medium improved fitness of a ridA mutant in coculture with wild type, making them near equal (Figure 3). Importantly, a minor, but statistically significant difference in fitness remained. These data supported, but were not conclusive, that the competitive defect of a ridA mutant was caused by 2AA-dependent damage to the target enzymes GlyA and IscS. The slight fitness defect that remained left the possibility that either i) decreased flux to 2-ketobutyrate (from 2AC) in the isoleucine biosynthetic pathway contributes to the fitness defect of a ridA mutant, or ii) 2AA-dependent damage to PLP-dependent target enzymes beyond GlyA and IscS contribute to the fitness defect of a ridA mutant in S. enterica.
RidA proteins are required for full fitness of E. coli
Past studies have identified multiple differences between the metabolic networks of S. enterica and E. coli 21, 33, 34. E. coli encodes two RidA proteins, RidA and TdcF, which have been assessed for a role in 2AA stress 21. The ridA gene is monocistronic, while tdcF is in the tdc operon that encodes enzymes involved in a putative threonine catabolic pathway 35, 36 and are thought to be expressed only under anaerobic conditions 37, 38. Evidence of 2AA stress was observed in E. coli only with significant manipulation of the genome and growth medium. Loss of TdcF and/or RidA had no observable effect on growth nor activity of known 2AA target enzymes, unless 2AA stress was artificially intensified by expression of a feedback-insensitive variant of IlvA (encoded by the ilvA219 allele) in the presence of exogenous serine 21.
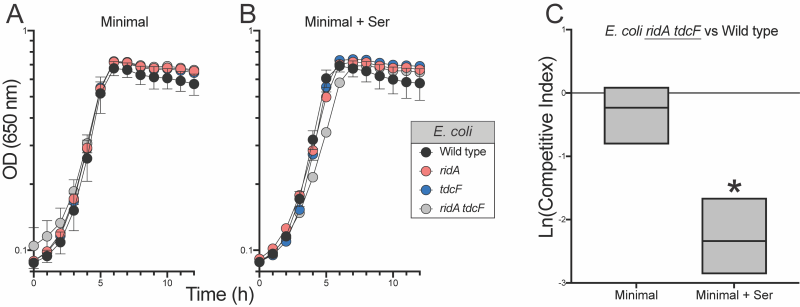
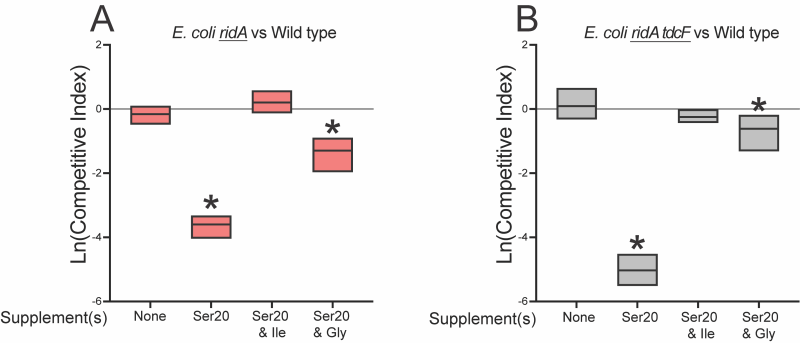
Unlike S. enterica, the absence of RidA proteins in E. coli (RidA and TdcF) had no effect on the growth, or competitive fitness of strains grown in minimal medium. In medium supplemented with 5 mM serine, the ridA tdcF double mutant strain had a small, but statistically significant, difference in growth rate (Figure 4) reminiscent of S. enterica ridA mutants grown in minimal glucose medium. Despite the minimal growth difference in monoculture, the ridA tdcF mutant was notably less fit than wild type in minimal medium supplemented with serine (Figure 4). Unlike the situation in S. enterica, there was no significant defect in IlvE activity of the E. coli tdcB ridA mutant when grown under these conditions that produced a clear fitness defect (5 mM serine) (Table 1).
–
Table 1. 2AA stress is not detectable in E. coli strains lacking RidA proteins.
|
|
Branched Chain Amniotransferase Activity* |
|||
|---|---|---|---|---|
|
Strain |
Relevant Genotype |
Minimal |
Minimal + Ser |
Minimal + Ser + Ile |
|
DM14520 |
Wild type |
130 ± 6 |
132 ± 4 |
243 ± 6 |
|
DM14597 |
ridA |
140 ± 20 |
110 ± 4 |
234 ± 23 |
|
DM14699 |
tdcF |
163 ± 15 |
163 ± 5 |
265 ± 22 |
|
DM14682 |
ridA tdcF |
131 ± 9 |
134 ± 7 |
220 ± 4 |
*Activity of Branched Chain Aminotransferase (IlvE) is reported as nmol 2KMV/mg protein.
E. coli strains were grown in minimal glucose medium with the addition of serine (5 mM) with or without isoleucine (1 mM) as indicated. The activity of IlvE was measured as described in Experimental Procedures. Data shown are the mean and standard error of two biological replicates assayed in three technical replicates. There was no significant difference in IlvE activity between wild-type and mutant strains grown in the same medium, or between growth in minimal with and without serine addition. IlvE activity was significantly higher when grown in the presence of serine and isoleucine than the same strain in the other media tested.
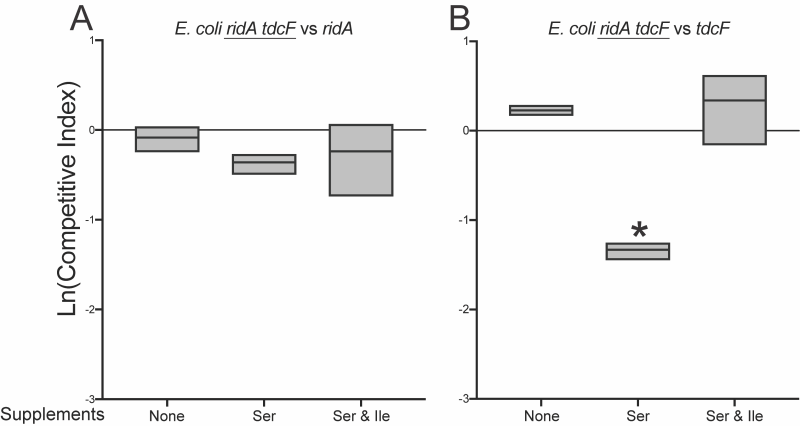
Additional competition experiments showed that RidA was the homolog critical for full fitness of E. coli grown in minimal medium with 5 mM exogenous serine (Figure 5). There was no difference between the fitness of the ridA tdcF double mutant and a ridA mutant. In contrast, the ridA tdcF double mutant was less fit than a tdcF mutant when grown in coculture. Isoleucine restored full fitness to the E. coli strains, eliminating the effect of the ridA mutation as found in S. enterica. These data were consistent with a scenario that 2AA stress negatively impacted fitness of E. coli.
–
2AA stress is induced in E. coli with increased serine concentration
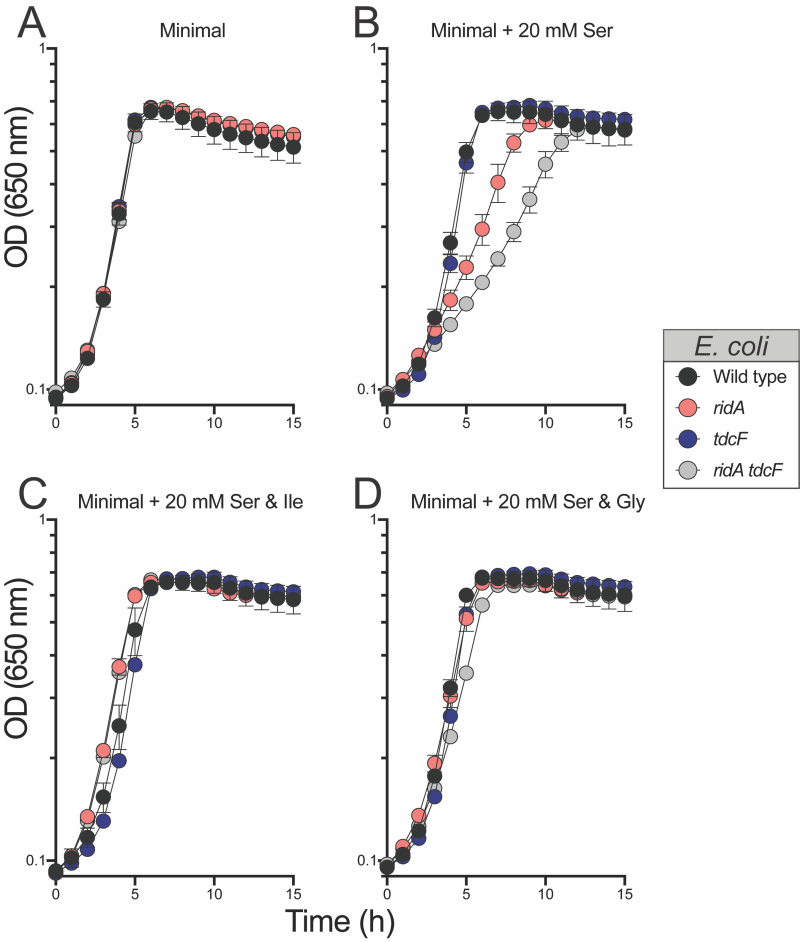
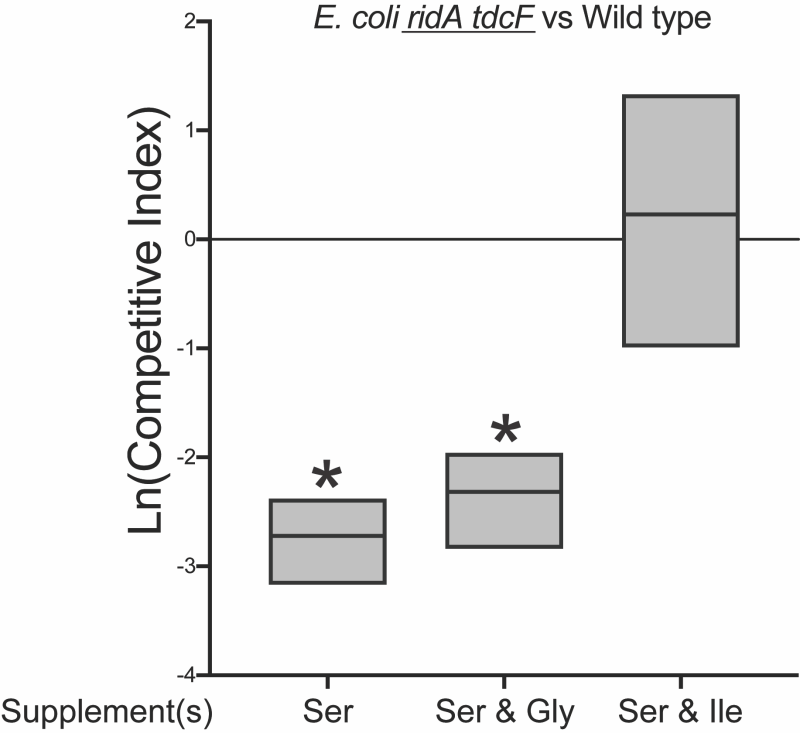
The above results could be explained in a scenario where 5 mM serine generated enough 2AA to compromise fitness, but not enough to cause detectable damage to target enzyme IlvE. If this was the case, increased concentrations of serine might increase 2AA production to a level that could generate a growth defect in monoculture. Consistently, both the ridA and ridA tdcF mutants had a growth defect with 20 mM serine, while the growth of wild type was not affected (Figure 6). The serine-induced growth defects of the ridA and ridA tdcF mutants were eliminated with the addition of isoleucine or glycine, consistent with the pattern in S. enterica with 2AA-dependent growth defects. As expected based on the growth defects in monoculture, E. coli ridA and ridA tdcF mutants were outcompeted by wild type in minimal medium containing 20 mM serine, and the ridA tdcF mutant having a more severe defect than the ridA single mutant (Figure 8). The addition of isoleucine restored full fitness to each stain in the presence of 20 mM serine. The observation that TdcF had an impact on fitness in the presence of elevated serine concentration was not entirely unexpected. E. coli strains with synthetically increased 2AA levels exhibited a growth defect only when both TdcF and RidA were absent, consistent with their redundant activities 21. In total, the results with E. coli suggested that either i) 2AA levels are generally lower in E. coli than S. enterica when strains lack RidA proteins, and/or ii) 2AA is less detrimental to the metabolic network of E. coli than it is to that of S. enterica. Neither of these directly addressed the disconnect between the lack of detectable 2AA damage (i.e., IlvE activity) and competitive fitness defect of an E. coli ridA mutant in minimal medium with 5 mM serine.
–
RidA has a role beyond 2AA stress control in E. coli
The data gathered for ridA mutants of E. coli appeared consistent with a model in which, i) the fitness defect on 5 mM serine is due to 2AA stress, despite the stress being too low to impair IlvE activity, ii) high levels of exogenous serine (20 mM) generate enough 2AA to cause a growth defect, and iii) glycine and isoleucine restore growth via the mechanism(s) characterized in S. enterica ridA mutants. Based on this scenario, glycine was expected to suppress the competitive fitness defect of the ridA tdcF mutant. Unexpectedly, glycine failed to improve fitness and the ridA tdcF mutant remained less fit than wild type in medium with serine (5 mM) and glycine (Figure 7). Furthermore, the addition of glycine did not restore competitive fitness defects of ridA or ridA tdcF mutants in the presence of 20 mM serine, despite its ability to restore growth in monoculture (Figure 8). A possible explanation for these results is that, in E. coli RidA has a role in optimizing flux through isoleucine biosynthesis. In this scenario, isoleucine contributes to restoring fitness by satisfying a need for isoleucine created by sluggish de novo synthesis, in addition to its role in decreasing 2AA generation. The finding that glycine eliminates the 2AA-generated growth defect in monoculture, while failing to restore the fitness defect of the ridA tdcF mutant, supports the conclusion that decreased fitness of E. coli on minimal medium with serine is not solely due to 2AA stress.
–
The kinetics of ECIlvA model 2AA and 2AC production
The above data suggested that fitness defects of E. coli ridA and ridA tdcF mutants in coculture were not solely due to 2AA-mediated damage to PLP-dependent enzymes. The deaminase activities of other members of the Rid protein family improve flux through their respective pathway(s) and thus increase fitness. Based on this precedent, it was considered possible that fitness defects of E. coli mutant strains in coculture were due to impaired flux to isoleucine in the absence of RidA. For this to be the case, the data indicated flux was reduced only when exogenous serine was present. In principle, impaired flux could be due to i) decreased 2AC formation by IlvA, or ii) decreased deamination of 2AC to 2KB in the absence of RidA. Only the former possibility appeared to be amenable to an effect by serine, since competition between serine and threonine for IlvA could potentially alter the 2AC:2AA ratio. In S. enterica, kinetic models indicated that the production of 2AC from threonine was not affected by the level of serine present in vivo 39. The catalytic parameters (KM and kcat) of the E. coli protein (ECIlvA) were determined with serine and threonine as substrates (Figure S1). Using these parameters of ECIlvA to model endogenous 2AC and 2AA production in E. coli indicated that 2AC production was not impacted by the increased production of 2AA (Table 2). The ratios of 2AC:2AA predicted by modeling were consistent between S. enterica and E. coli. In total, these data show that while 2AC genertion is not predicted to differ, the amount of 2AA relative to 2AC increases dramatically in E. coli strains grown with serine. In total the data are consistent with a scenario in which 2AA stress is not the sole cause of fitness defects in strains lacking RidA homologs. Fitness defects may be exacerbated by impaired flux through isoleucine biosynthesis that results from lower 2AC deamination in the absence of RidA activity, the direct effect of serine on this flux remains unclear.
–
Table 2. Modeled rates of product formation byECIlvA.
|
[Serine] (µM) |
[Threonine] (µM) |
2AA formation (µM/s) |
2AC formation (µM/s) |
|---|---|---|---|
|
68 |
180 |
6 |
84 |
|
5000 |
180 |
398 |
81 |
|
20000 |
180 |
1410 |
83 |
Modeled rates of endogenous product formation by ECIlvA using varied concentrations of serine and threonine. Data were calculated with an IlvA active site concentration of 10 mM, along with relevant kcat and KM values for each substrate (Figure S1) using the formula, and the substrate concentrations described in the Materials and Methods section. Higher serine concentrations were used assuming that the exogeneous concentration was equaled endogenously.
DISCUSSION
In S. enterica, ridA is one of many genes that do not generate an observable, or easily quantifiable, phenotype unless perturbations are applied to the system. In this study, we used cocultures to exacerbate minor growth defects and generate a quantifiable fitness phenotype for ridA mutants. Unlike canonical competition analyses that define fitness in terms competing for a single limiting resource, coculture experiments in this study defined fitness of a mutant compared to wild type for overall growth (requiring synthesis of all cellular components) in a defined minimal medium. This approach provided a sensitive assay for the detrimental effects of 2AA that were not detected during growth in monoculture.
In S. enterica, the requirement for RidA is revealed by adding serine to the growth medium. The presence of serine increases IlvA-mediated formation of the reactive enamine, 2AA. In the absence of added serine, 2AA stress in a ridA mutant is evidenced by decreased IlvE activity. Data herein allow the conclusion that 2AA stress decreases fitness of a S. enterica ridA mutant in minimal medium, despite the full growth of the mutants in monoculture. The restoration of fitness by isoleucine or glycine and thiamine supported this conclusion but did not eliminate the possibility of additional metabolic complexity.
Results with E. coli suggested a more nuanced situation than the 2AA stress paradigm defined with S. enterica. First, E. coli strains lacking RidA proteins had a significant fitness defect under conditions (i.e., minimal medium with 5 mM serine) where there was no quantifiable evidence of 2AA stress – i.e., lowered IlvE activity. Secondly, while the fitness defect was eliminated by the presence of isoleucine, glycine had little effect, further distinguishing the behavior of E. coli from S. enterica. Importantly, glycine restored growth of the ridA tdcF mutants of E. coli when 2AA levels were increased with elevated (20 mM) serine. Thus, if the competitive defect of the ridA tdcF mutant was caused by 2AA stress alone, glycine might be expected to completely restore fitness in coculture, which it did not. In total, the data suggest that the role of RidA proteins, at least in E. coli, extends beyond preventing accumulation of 2AA. Specifically, results were consistent with a scenario in which RidA has an additional role of increasing flux through de novo synthesis of isoleucine.
The experiments with E. coli suggested an expanded role for RidA, while raising additional questions about the structure of the metabolic network. Results of competition experiments suggested that RidA was needed to optimize flux to isoleucine in the presence of serine, but not in minimal medium. The kinetic parameters of ECIlvA, much like that of SEIlvA 39, suggest 5 mM serine increases the amount of 2AA without affecting the amount of 2AC generated in the cell. Therefore, the need for RidA to contribute to biosynthetic flux only in the presence of serine seems to be due to the ratio of 2AA/2AC, though the precise mechanism remains unclear. The lack of a quantifiable phenotype indicates the increase in 2AA mediated by 5 mM serine does not cause detectable damage to target enzymes in E. coli. It is possible that a higher ratio of 2AA:2AC amplifies a need for RidA to generate 2-ketobutyrate at a rate that generates isoleucine efficiently. Such efficiency appears to be important only when in coculture with a strain fully proficient at isoleucine synthesis, due to the presence of RidA. Consider that 2AC has three fates in the E. coli cell, two of which lead to isoleucine, and one contributes to the synthesis of thiamine via TrpD forming phosphoribosylamine (PRA) from 2AC and phosphoribosylpyrophosphate (PRPP) 8 (Figure 9). The metabolic node encompassing 2AC and PRA is structured differently in S. enterica and E. coli, consistent with a difference in the role of RidA in the two organisms 6, 9, 33, 40.
–
It is worth noting that a previous study increased the accumulation of 2AA in E. coli by utilizing both serine and a variant of IlvA that was insensitive to allosteric inhibition by isoleucine 21. Under these conditions, 2AA stress that compromised growth in E. coli in monoculture was generated, and glycine failed to restore growth. This result apprears inconsistent with the scenario proposed here. However, IlvA is expected to be critical in generating appropriate ratios of reactive intermediates (i.e., 2AA and 2AC) which, in turn, can influence flux through numerous metabolic pathways. Because of this connection, eliminating allosteric regulation of IlvA will have consequences beyond the specific accumulation of 2AA, offering an explanation for the differences found in the two studies.
This study, like others that have dissected metabolic integration, emphasizes the significance of metabolic network structure in defining the response of each organism to perturbation. The response of an organism to metabolic network perturbations must be defined on a case-by-case basis. This realization highlights the fact that metabolism is a complex system that has been finely tuned by evolution, rather than a network assembled fortuitously from components that evolved for individual efficacy. Specific to this work, E. coli experiences less 2AA accumulation and/or the metabolic network is more resistant to the resulting damage, while the synthesis of isoleucine in this organism can be a limiting factor under some growth conditions. In contrast, S. enterica is sensitive to 2AA accumulation, yet the biosynthesis of isoleucine appears to be sufficient for fitness even in the absence of RidA. These results reflect differential responses of two metabolic networks to perturbation, in spite of the similarity of individual components. Defining the consequences of perturbation in multiple systems (i.e., organisms) will continue to expand our knowledge of the breadth of strategies that are germane when modeling metabolism.
MATERIALS AND METHODS
Strains, media, and chemicals
Strains used in this study are derivatives of Salmonella enterica serovar Typhimurium LT2 or Escherichia coli K-12 and are listed in Table 3. All strains were grown at 37°C unless otherwise stated. S. enterica strains were routinely grown in Nutrient Broth (NB; Difco; 8 g/liter, 5 g/liter NaCl). E. coli strains were grown in Lysogeny Broth (LB; 10 g/liter tryptone, 5 g/liter NaCl, 5 g/liter yeast extract) as a rich medium. No-carbon E (NCE) medium with 11 mM glucose, supplemented with 0.1 mM MgSO4 and trace elements was used as a minimal medium 41. Other supplements were added as noted to the following concentrations: L-isoleucine (1 mM), L-glycine (1 mM), thiamine (100 nM), L-aspartate (1 mM), and L-serine (5 mM). Antibiotics were used as indicated: kanamycin (Km; 50 μg/ml for rich media or 12.5 μg/ml for minimal media) and chloramphenicol (Cm; 20 μg/ml for rich media or 5 μg/ml for minimal media). Chemicals were purchased from MilliporeSigma (St. Louis, MO).
Table 3. Strains used in this study.
|
Strain |
Genotype |
Source |
|---|---|---|
|
S. enterica |
||
|
DM3480 |
ridA3::MudJa |
Laboratory collection |
|
DM4748 |
ilvA595::Tn10 |
Laboratory collection |
|
DM6143 |
ridA3::MudJ ilvA595::Tn10 |
Laboratory collection |
|
DM9404 |
Wild type |
Laboratory collection |
|
DM17845 |
ridA4::Cm |
This study |
|
|
||
|
E. coli |
||
|
DM14520 |
E. coli K-12 MG1655 wild type (F– l– |
Laboratory collection |
|
DM14597 |
ridA790::Km |
Laboratory collection |
|
DM14669 |
tdcF12::Km |
Laboratory collection |
|
DM14670 |
tdcF13::Cm |
Laboratory collection |
|
DM14682 |
ΔridA890 |
Laboratory collection |
|
DM14820 |
ridA12::Cm |
Laboratory collection |
|
DM15051 |
ΔridA890 ΔtdcF14 |
Laboratory collection |
|
DM18490 |
E. coli BL21-AI/pTEV18-ilvA (K12) |
This study |
Growth analyses
Cultures were started from a single colony and grown overnight in rich medium with shaking at 37°C (2 ml). Cells were pelleted and resuspended in an equal volume of sterile saline. Cell suspensions (5 μl) were used to inoculate the indicated medium (195 μl). Growth was monitored in a 96-well plate as the change in optical density at 650 nm (OD650) over time using a BioTek ELx808 plate reader (BioTek Instruments, Winooski, VT) with a slow shaking speed (210 rpm). Data were plotted using Graphpad Prism version 8.4. Growth experiments were performed with three independent biological replicates unless otherwise stated.
Competition assays
Competition assays were performed generally as described 22, 44, 45. Briefly, overnight cultures of each strain were grown in rich medium (2 ml) at 37°C with shaking. Cells were pelleted and resuspended in 2 ml sterile saline and optical densities at 650 nm (OD650) were normalized to 0.1. 5 ml of the indicated medium was inoculated with 50 μl of cell suspension from each of the two competing strains. Control experiments were performed for each condition in which strains of the same relevant genotype, with different antibiotic resistance cassettes, were competed against each other. These controls accounted for differences in competitive fitness caused by the different antibiotic resistance cassettes in each strain. Colony forming units (CFUs) of cocultures were determined by spreading serial dilutions at T=0 (i.e., before incubation of the cocultures) onto media to differentiate between each respective strain. For S. enterica, media were as follows: (i) minimal medium supplemented with L-serine, selective for wild type (DM9404); (ii) minimal medium supplemented with chloramphenicol, selective for a chloramphenicol-resistant ridA mutant (DM17845), and (iii) minimal medium supplemented with kanamycin, selective for a kanamycin-resistant ridA mutant (DM3480). For E. coli, media were as follows: (i) LB supplemented with chloramphenicol, selective for chloramphenicol-resistant ridA and ridA tdcF mutants (DM14820, DM14682), and (ii) LB supplemented with kanamycin, selective for kanamycin-resistant ridA and tdcF mutants (DM14597, DM14669). Some E. coli strains (DM14520, DM15051) did not contain antibiotic resistance cassettes and could not be isolated from cocultures by spreading onto selective media. CFUs of these strains were determined by counting CFUs on LB media and subtracting the CFUs of the competing strain, which could be selected for with antibiotics. Cocultures were grown to stationary phase at 37°C with shaking. CFUs were enumerated for cultures after co-incubation as described above. The competitive index (CI) was defined and calculated as follows:
–
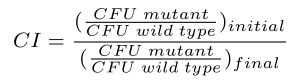 |
Each competition assay was performed at least three times with similar results. GraphPad Prism version 8.4 was used for data plotting and statistical analysis.
Protein purification
E. coli K12 ilvA was cloned into BspQI sites of pTEV18 as described previously. The resulting plasmid, pTEV18-ilvA (K12), was electroporated into BL21-AI. The resulting strain (DM18490) was grown overnight in 40 ml of Terrific Broth (TB; 12 g/liter tryptone, 24 g/liter yeast extract, 0.4% glycerol (v/v)) supplemented with 150 mg/ml ampicillin. 15 ml of the overnight culture was used to inoculate each of two 2.8-liter baffle-bottomed Fernbach flasks containing 1.5 liters of TB with 150 mg/ml ampicillin. The resulting cultures were grown at 30°C with shaking at 135 rpm to an OD650 of 0.5 before the addition of arabinose (0.2% (w/v)) and IPTG (200 mM). Cultures were then grown at 23°C with shaking at 135 rpm for 18 h before being harvested by centrifugation at 6000 x g for 20 min. Cell pellets were resuspended in 2 ml/g cells of lysis buffer (50 mM potassium phosphate, 150 mM NaCl, 100 mM PLP, 2 mg/ml lysozyme, 0.2 mg/ml DNase, pH 7.5) and incubated on ice for 45 min. Cells were mechanically lysed using a Constant Systems OneShot Cell Disruptor at 20 kpsi. The lysate was then clarified by centrifugation at 45000 x g for 30 min before being filtered using a 0.45 mm syringe-driven filter. Filtered lysate was then loaded onto a 5 ml HisTrap HP column (GE Healthcare). The column was washed with 3 column volumes (CV) of 100% Binding Buffer (50 mM potassium phosphate, 150 mM NaCl, 20 mM imidazole, pH 7.5), followed by 3 CV of 96% Binding Buffer and 4% Elution Buffer (50 mM potassium phosphate, 150 mM NaCl, 500 mM imidazole, pH 7.5). IlvA was eluted with a linear gradient from 4-100% Elution Buffer over the course of 10 CV. A flow rate of 2 ml/min was used to each step in protein purification. Fractions containing eluted protein were assessed for purity before being concentrated using an Amicon Ultra-15 centrifugal filter (MilliporeSigma) with a molecular weight cutoff of 30 kDa. Buffer exchange of the protein into Storage Buffer (50 mM potassium phosphate, 150 mM NaCl, 10% glycerol (v/v) pH 7.5) was performed using a PD-10 column (Cytiva). Protein concentration was determined using the Peirce BCA Protein Assay Kit (Thermo Scientific). Purified IlvA was drop frozen in liquid nitrogen and stored at -80°C until use. Purity of the protein was determined by densitometer tracing to be 94% (Figure S2).
Enzyme assays
All enzyme assays were performed at room temperature.
Transaminase B assay-cell extract
Cultures were grown overnight in LB (2 ml) at 37°C with shaking, and cells were pelleted and resuspended in an equal volume of sterile saline. Cell suspensions (125 μl) were used to inoculate 5 ml of the relevant medium and the resulting cultures were grown to stationary phase. Cells were then pelleted, resuspended in 10 ml of NCE medium before being pelleted and frozen at -80oC until use. Cell pellets were thawed and resuspended in 1 ml of 50 mM potassium phosphate (pH 7.5) and lysed mechanically with a Constant Systems Limited One Shot (United Kingdom) at 20 kpsi. Cell lysate was clarified by centrifugation at 17000 x g for 20 min at 4°C.
Transaminase B activity was assayed as described previously 46. Briefly, cell free extract (50 μl) was added to a reaction mixture containing α-ketoglutarate (10 mM) in potassium phosphate buffer (50 mM, pH 7.5). The reaction was initiated with the addition of L-isoleucine (to 20 mM) and incubated at 37C for 20 minutes. Resulting 2-ketomethylvalerate (2KMV) was derivatized by 2,4-dinitrophenylhydrazine (DNPH, 200 μl) to yield a hydrazone, which was treated with 0.5 N HCl. The resulting organic layer was removed and mixed with 1.5 N NaOH to allow formation of an aqueous layer containing a chromophore with an absorbance at 540 nm. The aqueous layer was extracted, and absorbance was measured using a SpectraMax M2 microplate reader (Molecular Devices). 2KMV concentration was quantified by comparing to a standard curve, which was generated by monitoring the absorbance of reaction mixtures with known 2KMV concentrations that were subjected to the same incubation and extraction steps as described above for the cell free extracts. The protein concentration of each lysate was determined using the bicinchoninic acid (BCA) assay (Pierce). Activity is reported as nanomoles of 2KMV per milligram of protein per cell lysate. Data plotting and statistical analyses were conducted using GraphPad Prism version 8.4.
Serine/Threonine Dehydratase assay -purified protein
The rates of 2-ketobutyrate and pyruvate formation were determined by measuring the absorbance at 230 nm39, 3, 47 every 10 s for 2 min. Reactions were carried out in a quartz 96-well plate and measurements were acquired using a SpectraMax Plus spectrophotometer (Molecular Devices). Reaction mixtures (100 ml) contained 50 mM potassium phosphate buffer (pH 7.5), 200 nM of purified IlvA. Reactions were initiated with the addition of the indicated concentrations of L-threonine or L-serine. The absorbance at 230 nm of known concentrations of either 2-ketobutyrate or pyruvate were used to generate a standard curve to convert the change in absorbance (DA230/min) to the rate of product formation (mmol product/min) for each reaction. Data visualization and statistical analyses were performed using GraphPad Prism version 8.4.
The rates of 2AA and 2AC synthesis were modeled using the equation below 39. Vprod refers to the rate of product formation (2AA or 2AC) in µM/s, kcat(Sub) refers to the catalytic constant for ECIlvA using the relevant substrate (s-1), [Et] is the active site concentration of IlvA in E. coli, [Sub] is the concentration of substrate for ECIlvA in vivo in µM, [Comp Sub] is the concentration of competing substrate in µM, KM(Sub) is the Michaelis-Menten constant for ECIlvA using the relevant substrate in mM, and KM(Comp Sub) is the Michaelis-Menten constant for ECIlvA using the relevant competing substrate in mM. When modeling the rate of 2AC formation, “Sub” refers to L-threonine and “Comp Sub” refers to L-serine. When modeling the rate of 2AA formation, “Sub” refers to L-serine and “Comp Sub” refers to L-threonine. 10 µM was used as the value for active site concentrations for ECIlvA in vivo 48 and physiological concentrations of L-serine (68 µM) and L-threonine (180 µM) in E. coli 49, as well as elevated concentrations of L-serine (5000 µM and 20000 µM) were used when calculating the product formation by ECIlvA.
–
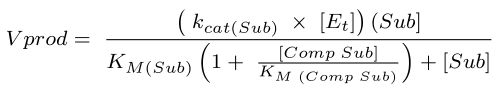 |
REFERENCES
For full bibliography please see the pdf file.
SUPPLEMENTAL INFORMATION
![]() Download Supplemental Information
Download Supplemental Information
ACKNOWLEDGMENTS
This work was supported by funding from the NIH (GM095837 and GM153189) to D.M.D. and (T32GM007103) to R.L.F.
COPYRIGHT
© 2024

RidA proteins contribute to fitness of S. enterica and E. coli by reducing 2AA stress and moderating flux to isoleucine biosynthesis by Fulton et al. is licensed under a Creative Commons Attribution 4.0 International License.