Back to article: RidA proteins contribute to fitness of S. enterica and E. coli by reducing 2AA stress and moderating flux to isoleucine biosynthesis
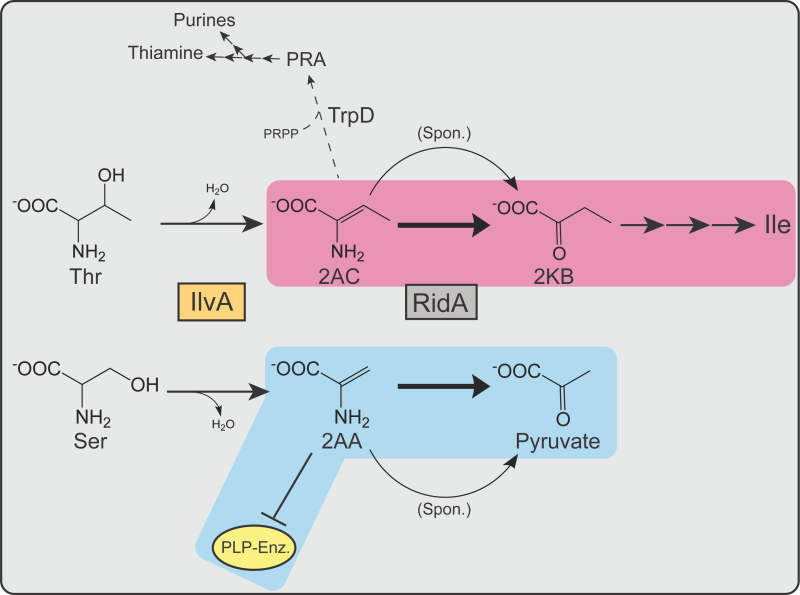
FIGURE 9: RidA proteins contribute to fitness in S. enterica and E. coli by moderating flux through different metabolic nodes. Shown are the pathways relevant to RidA activity in S. enterica and E. coli. IlvA (orange box) dehydrates serine or threonine, generating 2AA or 2AC, respectively. 2AA and 2AC are spontaneously deaminated by water (curved arrows) or, more rapidly, by RidA proteins (grey box, bold arrows) to the corresponding ketoacid product – pyruvate or 2KB. 2KB serves as a substrate for Ile synthesis. 2AA can irreversibly damage PLP-dependent enzymes (yellow oval) in the absence of RidA. Metabolic nodes in which RidA activity primarily contributes to fitness of each organism are highlighted in blue (S. enterica) or magenta (E. coli). 2AC, along with PRPP, is also used as a substrate by TrpD (dashed arrow) to generate PRA, a precursor to purines and thiamine.