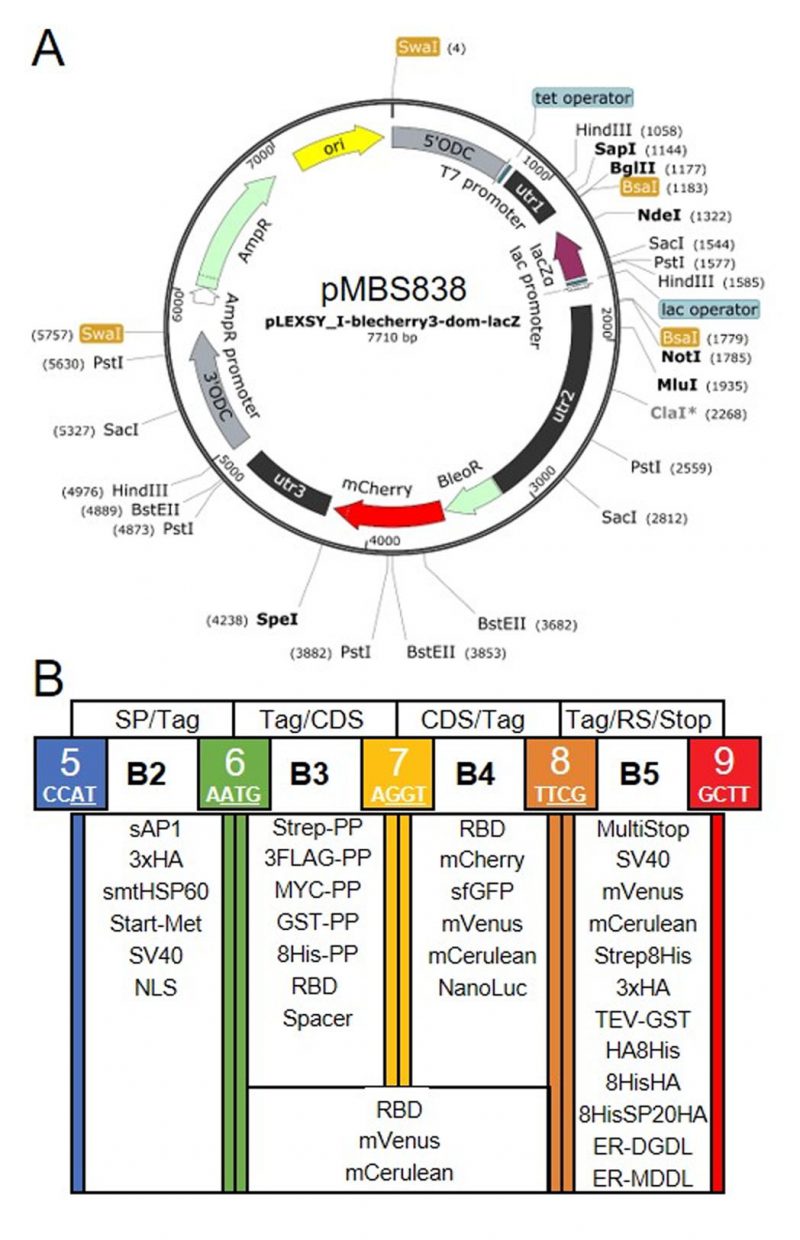
FIGURE 1: Genetic parts of the L. tarentolae MoClo kit. (A) Map of the destination vector for the L. tarentolae MoClo kit based on the pLEXSY_I-blecherry3 plasmid from Jena Bioscience. BsaI restriction sites were removed and the coding region for lacZα flanked by BsaI restriction sites was introduced to yield CCAT and GCTT fusion sites upon digestion with BsaI. (B) List of 34 Level 0 MoClo parts for positions B2 to B5 that are compatible with the MoClo syntax for plants and algae. The color code for fusion sites was adopted from the Chlamydomonas MoClo kit [35]. Nucleotides used in codons are underlined in white. SP, signal peptide; CDS, coding sequence; RS, retention signal; sAP1, secreted acid phosphatase 1; HA, human influenza hemagglutinin; Met, methionine; Strep, streptavidin; PP, PreScission protease cleavage site; GST, glutathione transferase; RBD, receptor binding domain of SARS-CoV-2; SV40, simian-virus 40; TEV, tobacco etch virus protease cleavage site; SP, serine-proline repeat; ER, endoplasmic reticulum.
- Crozet P, Navarro FJ, Willmund F, Mehrshahi P, Bakowski K, Lauersen KJ, Perez-Perez ME, Auroy P, Gorchs Rovira A, Sauret-Gueto S, Niemeyer J, Spaniol B, Theis J, Trosch R, Westrich LD, Vavitsas K, Baier T, Hubner W, de Carpentier F, Cassarini M, Danon A, Henri J, Marchand CH, de Mia M, Sarkissian K, Baulcombe DC, Peltier G, Crespo JL, Kruse O, Jensen PE, et al. (2018). Birth of a photosynthetic chassis: a MoClo toolkit enabling Synthetic Biology in the microalga Chlamydomonas reinhardtii. ACS Synth Biol 7(9): 2074-2086. 10.1021/acssynbio.8b00251