Research Reports:
Microbial Cell, Vol. 6, No. 10, pp. 482 - 490; doi: 10.15698/mic2019.10.694
Proline metabolism regulates replicative lifespan in the yeast Saccharomyces cerevisiae
1 Department of Frontier Bioscience, Faculty of Bioscience, Nagahama Institute of Bio-Science and Technology, 1266 Tamura, Nagahama, Shiga 526-0829, Japan.
2 Division of Biological Science, Graduate School of Science and Technology, Nara Institute of Science and Technology, 8916-5 Takayama, Ikoma, Nara 630-0192, Japan.
Keywords: proline, replicative lifespan, stress response, amino acid metabolism, yeast.
Abbreviations:
OPLS-DA – orthogonal projection to latent structure discriminant analysis,
PCA – principal component analysis,
WT – wild type.
Received originally: 29/04/2019 Received in revised form: 19/08/2019
Accepted: 16/09/2019
Published: 24/09/2019
Correspondence:
Yukio Mukai, Department of Frontier Bioscience, Faculty of Bioscience, Nagahama Institute of Bio-Science and Technology, 1266 Tamura, Nagahama, Shiga 526-0829, Japan; Tel.: +81-749-64-8163; Fax: +81-749-64-8140; y_mukai@nagahama-i-bio.ac.jp
Hiroshi Takagi, Division of Biological Science, Graduate School of Science and Technology, Nara Institute of Science and Technology, 8916-5 Takayama, Ikoma, Nara 630-0192, Japan; Tel.: +81-743-72-5420; Fax: +81-743-72-5429; hiro@bs.naist.jp
Conflict of interest statement: The authors declare no conflict of interest.
Please cite this article as: Yukio Mukai, Yuka Kamei, Xu Liu, Shan Jiang, Yukiko Sugimoto, Noreen Suliani binti Mat Nanyan, Daisuke Watanabe and Hiroshi Takagi (2019). Proline metabolism regulates replicative lifespan in the yeast Saccharomyces cerevisiae. Microbial Cell 6(10): 482-490. doi: 10.15698/mic2019.10.694
Abstract
In many plants and microorganisms, intracellular proline has a protective role against various stresses, including heat-shock, oxidation and osmolarity. Environmental stresses induce cellular senescence in a variety of eukaryotes. Here we showed that intracellular proline regulates the replicative lifespan in the budding yeast Saccharomyces cerevisiae. Deletion of the proline oxidase gene PUT1 and expression of the γ-glutamate kinase mutant gene PRO1-I150T that is less sensitive to feedback inhibition accumulated proline and extended the replicative lifespan of yeast cells. Inversely, disruption of the proline biosynthetic genes PRO1, PRO2, and CAR2 decreased stationary proline level and shortened the lifespan of yeast cells. Quadruple disruption of the proline transporter genes unexpectedly did not change intracellular proline levels and replicative lifespan. Overexpression of the stress-responsive transcription activator gene MSN2 reduced intracellular proline levels by inducing the expression of PUT1, resulting in a short lifespan. Thus, the intracellular proline levels at stationary phase was positively correlated with the replicative lifespan. Furthermore, multivariate analysis of amino acids in yeast mutants deficient in proline metabolism showed characteristic metabolic profiles coincident with longevity: acidic and basic amino acids and branched-chain amino acids positively contributed to the replicative lifespan. These results allude to proline metabolism having a physiological role in maintaining the lifespan of yeast cells.
INTRODUCTION
Microorganisms and plants are exposed to various environmental stresses during their lifespan and have developed a variety of adaptation strategies against these stresses (e.g., altered membrane composition, induction of stress-responsive proteins, and accumulation of compatible solutes) [1]. Proline is known to function in vitro as a stress protectant, namely a protein and membrane stabilizer, protein-folding chaperone, and reactive oxygen species (ROS) scavenger [2][3][4]. Proline also functions as a compatible solute, a small molecule whose presence in large numbers helps organisms survive extreme osmotic stress [1][5]. In the budding yeast Saccharomyces cerevisiae, the synthesis of proline from glutamate in the cytoplasm is catalyzed by three enzymes: Pro1p (γ-glutamyl kinase), Pro2p (γ-glutamyl phosphate reductase), and Pro3p (Δ1-pyrroline-5-carboxylate (P5C) reductase) [6][7]. Proline is also synthesized from arginine by two enzymes: Car1p (arginase) and Car2p (ornithine transaminase) [8]. On the other hand, excess proline is degraded to glutamate in mitochondria by two enzymes, Put1p (proline oxidase) and Put2p (P5C dehydrogenase), and inhibits the enzymatic activity of Pro1p by negative feedback inhibition [9]. Unlike bacterial and plant cells, yeast cells do not elevate proline levels in response to various stresses [10]. To date, however, we have found that the Asp154Asn and Ile150Thr variants of Pro1p were less sensitive to feedback inhibition, leading to excess proline synthesis [11][12]. Thus, S. cerevisiae cells accumulated proline by expressing the PRO1-D154N and PRO1-I150T genes encoding the above variants and by disrupting the PUT1 gene encoding proline oxidase [13]. Proline accumulation was shown to confer stress tolerance to freezing, desiccation, oxidation, and ethanol on yeast cells [11][13][14][15][16][17][18][19].
–
Numerous genes involved in determining lifespan have been identified in a variety of model organisms including yeast [20][21][22][23]. The replicative lifespan of a yeast cell is defined as the number of daughter cells produced from a mother cell before dying [24] and may be similar to the aging of mitotically active cells in multicellular organisms [25][26]. In addition to metabolic enzymes [27][28][29][30][31][32], it has also been shown that stress response factors are implicated in the replicative lifespan: e.g., protein kinase Sch9p and transcription factors Msn2p and Msn4p [28][33]. Sch9p is involved in the transactivation of osmostress-responsive genes and is required for TORC1-mediated regulation of ribosomal biogenesis and translation initiation through phosphorylation by Tor1p kinase [34]. Msn2p/Msn4p are transcriptional activators in response to various stress conditions [35][36].
–
Since proline functions as a stress protectant in yeast, as described above, we wondered whether intracellular proline is involved in the yeast replicative lifespan. Accordingly, the replicative lifespan of yeast mutants with altered proline content was measured and the data was statistically correlated to intracellular proline and other amino acids.
RESULTS AND DISCUSSION
Extended replicative lifespan observed in yeast cells with increased intracellular proline content
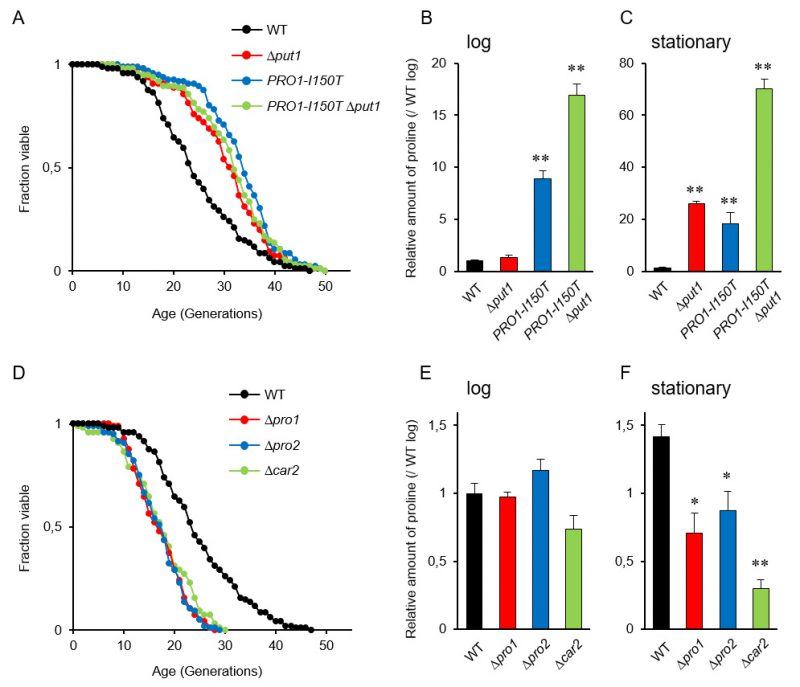
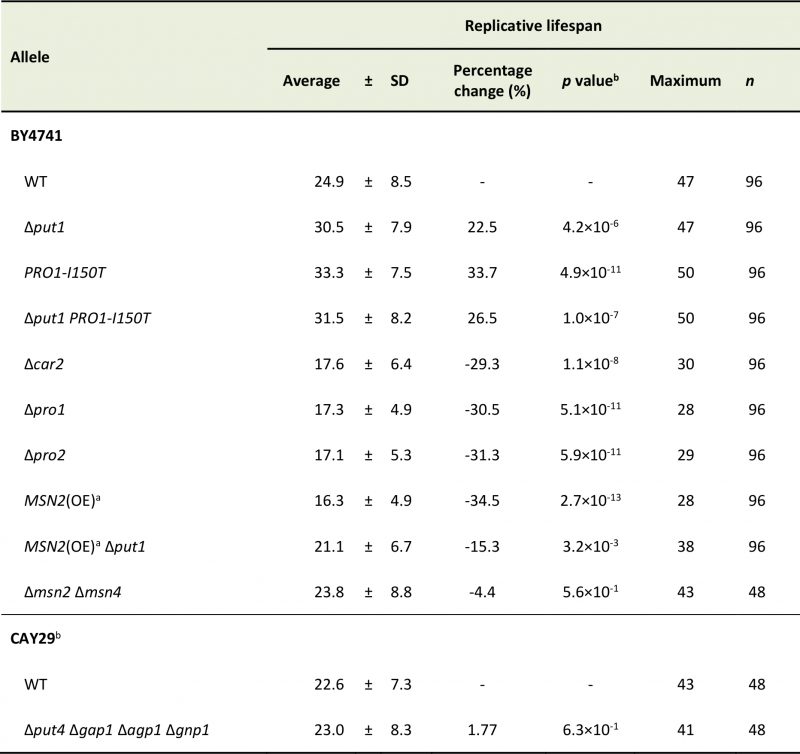
It is known that accumulation of proline in yeast cells confers tolerance to various stresses, e.g., freezing, desiccation, oxidation, and ethanol [9], and that several stress-resistant mutants are long lived [28]. Accordingly, we expected that proline accumulation would extend the replicative lifespan of yeast cells. The deletion mutant of PUT1, encoding proline oxidase, and a mutant expressing PRO1-I150T, which encodes the γ-glutamyl kinase variant (Ile150Thr) with desensitization to feedback inhibition by proline [12], exhibited markedly longer lifespans on YPD medium compared with the wild-type strain (WT) (Figure 1A; replicative lifespan measurements from this study are shown in Table 1). The Δput1 PRO1-I150T double-mutant did not show further lifespan extension compared with each single mutant.
–
–
We confirmed the proline accumulation in the Δput1 and PRO1-I150T mutants. As previously reported [18], the proline level of the Δput1 mutant during logarithmic growth in YPD medium was comparable to that of WT, while that of the PRO1-I150T mutant was 9-fold higher than that of WT (Figure 1B). In the stationary phase (after three days of cultivation), both the Δput1 and PRO1-I150T mutant strains accumulated high levels of proline compared with WT (Figure 1C). The proline levels of WT cells in the stationary phase were almost the same (1.4-fold) as those in the log phase. These results suggest that an increase in proline levels extends the replicative lifespan of yeast cells when grown aerobically in YPD medium. In addition, even though the Δput1 PRO1–I150T double-mutant lifespan was similar to that of each single mutant, the double-mutant strain highly accumulated proline in the log and stationary phases (17- and 70-fold, respectively, higher than WT in each phase). These results indicate that excess proline does not lead to further lifespan extension.
–
In the Δput1 mutant, proline is not accumulated in the log phase probably due to high consumption of proline for cell growth. In contrast, cell proliferation will stop in the stationary phase, leading to proline accumulation that could delay the replicative aging, consequently extending lifespan. This speculation might be related to our previous results that the stationary phase-induced genes were mostly up-regulated in aged cells [38].
–
Shortened replicative lifespan observed in yeast cells with reduced intracellular proline content
Since the accumulation of proline extended the replicative lifespan of yeast cells, we assumed that a decrease of intracellular proline levels would shorten its lifespan. To address this assumption, we examined the effect of deleting three genes involved in proline biosynthesis on the replicative lifespan. The PRO1 and PRO2 genes encode the first and second enzymes, respectively, in proline biosynthesis from glutamate [6][7]. The CAR2 gene encodes the second enzyme in arginine degradation to supply glutamate-γ-semialdehyde as a proline precursor [8]. As expected, deletion of PRO1, PRO2, or CAR2 markedly shortened the lifespan on YPD medium (Figure 1D). Intracellular proline levels in the Δpro1, Δpro2, and Δcar2 mutants were comparable to those in WT when these cells were cultivated to the log phase (Figure 1E). However, when cultivated to the stationary phase, all the mutants had significantly lower level of proline compared with that of WT (Figure 1F). These results suggest that a decrease in the proline levels shortens the replicative lifespan of yeast cells when grown aerobically in YPD medium.
–
It has been reported that S. cerevisiae has four proline transporter genes: two general amino acid permease genes, GAP1 and AGP1, the proline permease gene PUT4, and the glutamine permease GNP1 gene [37]. Hence, we speculated that a deficiency in proline permease activity would decrease the intracellular proline levels, leading to a shortened lifespan. However, the quadruple deletion of the proline transporter genes did not change the lifespan of yeast cells on YPD medium (Figure S1A). This result might be reasonable because the proline level in the quadruple mutant seemed to be comparable to the level of WT (Figure S1B). Therefore, glutamate taken up from YPD medium might be utilized to proline synthesis in the cell even when the proline transporter genes are deficient.
–
The stress-responsive transcription activator genes MSN2/MSN4 are involved in replicative lifespan
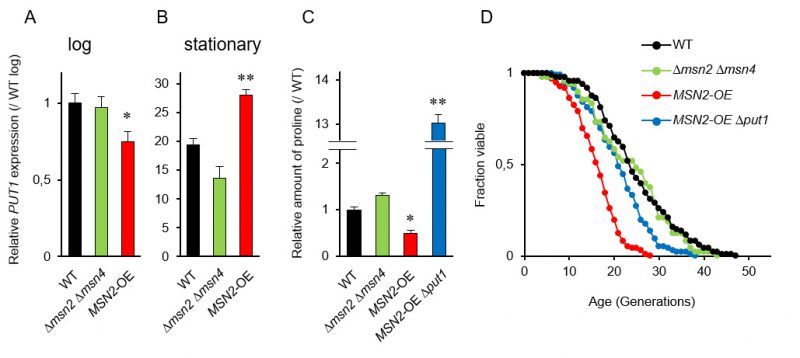
Since the promoter region in PUT1 contains two stress response elements (STREs), which are bound by two paralogous transcriptional activators Msn2p and Msn4p [36], we postulated that MSN2 and MSN4 regulate the replicative lifespan by mediating the proline levels. RT-qPCR analysis showed that the induction of PUT1 transcription in WT in the stationary phase was about 20-fold greater than that in the log phase (Figure 2A and 2B). In the stationary phase, overexpression of MSN2 driven by the TDH3 promoter significantly enhanced the PUT1 transcription and decreased proline level (Figure 2B and 2C). On the other hand, when MSN2 and MSN4 were simultaneously deleted, there were no significant changes in the transcript level of PUT1 and the proline level compared to those of WT. Thus, the Δmsn2 Δmsn4 double-mutant exhibited a normal replicative lifespan (Figure 2D). Overexpression of MSN2 remarkably shortened the replicative lifespan, which was restored almost to the lifespan of WT by deletion of PUT1, although the proline level in the MSN2-overexpressing and put1-deleted strain was 13-fold higher than that in WT (Figure 2C and 2D). These results support our hypothesis that intracellular proline, in part, modulates the replicative lifespan.
–
–
To verify the above results that intracellular proline levels, which significantly change in the stationary phase, might regulate longevity, we performed correlation analysis between the proline levels in the stationary phase and the mean lifespan. The correlation coefficient (R=0.687) suggests that intracellular proline positively regulates longevity of yeast cells (Figure S2).
–
The transcript level of PUT1, when driven by the constitutive TDH3 promoter, was not enhanced in the log phase. This result suggests that the stability of the PUT1 transcripts depends on the growth phase. We previously observed that many genes that are induced during the stationary phase are transactivated at the early stages of replicative senescence of yeast cells [38]. Similarly, the mRNA levels of PUT1 were also increased in 11th generation cells compared with 1st generation cells [38]. Therefore, the age-dependent expression of PUT1 suggests that the control of intracellular proline levels is important for yeast cells during senescence. The stress-responsive transcription activator Msn2p, potentially also Msn4p, might be activated at the early senescence stage to upregulate the senescence-specific genes including PUT1, resulting in maintaining the lifespan of yeast cells.
–
Amino acid analysis of yeast mutants involved in proline metabolism
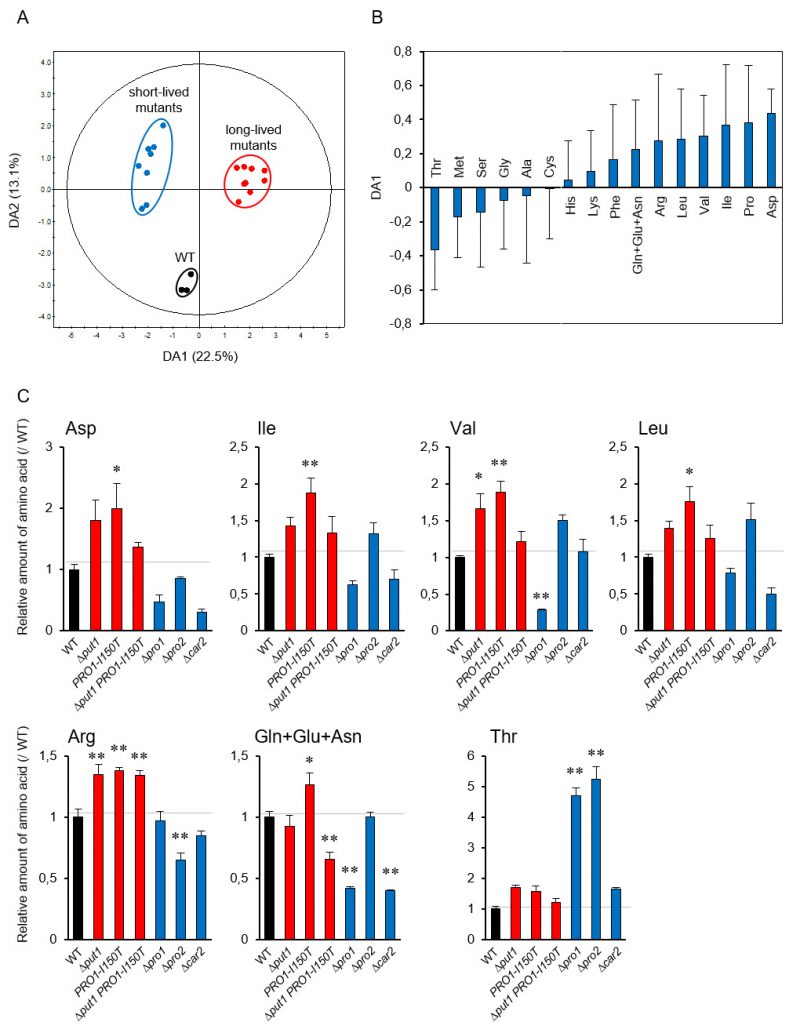
To determine the metabolic effects of increasing and decreasing intracellular proline levels, we analyzed the amino acid contents in the yeast strains used in this study in the log and stationary phases (Figure S3). In WT, half of the amino acids were decreased in the stationary phase relative to the log phase, whereas leucine and valine contents were increased. Thus, the changes took place in the stationary phase, but proline was unique in showing a correlation to the lifespan.
–
To link metabolism of proline to individual amino acids [38], we performed a principal component analysis (PCA) using the amino acid contents in the yeast mutants involved in proline metabolism to investigate the lifespan-dependent amino acid metabolism. The PCA score plot did not indicate the expected separation of these strains in the log and stationary phases (Figure S4A and S4B). Accordingly, to identify which amino acids contribute to the lifespan determination, we performed orthogonal projection to latent structure discriminant analysis (OPLS-DA) using the above amino acid dataset. The OPLS-DA score plot indicates a clear separation between WT and the short- and long-lived mutants (Figure 3A). This separation depended on lifespan along the DA1 axis. The loading plot for DA1 showed that, in addition to proline, acidic amino acids (aspartate and glutamate), branched-chain amino acids (isoleucine, valine, and leucine), and basic amino acids (arginine and lysine) positively contributed to longevity, whereas serine, threonine, and methionine were negatively involved (Figure 3B). These results were confirmed by the individual results (Figure 3C). Aspartate, isoleucine, valine, leucine, and arginine tended to be increased in the proline-increased mutants and decreased in the proline-decreased mutants. Threonine was remarkably increased in the proline-decreased mutants. These results suggest that the metabolic changes due to changing intracellular amino acid levels cause the lifespan determination.
–
–
In this study, we examined the phenotypes of various strains derived from BY4741 and CAY29 with S288C genetic background. It should be noted that genotype-phenotype outcomes are not always the same in all strains because of the influence of nuclear genome variation and that of the mitochondrial genome. To demonstrate our hypothesis that proline metabolism has a physiological role of influencing the replicative aging of yeast, it is necessary to construct other strains with different genetic background and to check their phenotypes. We initially focused on the involvement of proline in the ‘replicative' aging of yeast cells. From our results, it is intriguing whether proline accumulated at the stationary phase extends ‘chronological' lifespan of yeast cells.
–
Recently, higher proline levels were found to increase longevity in Caenorhabditis elegans [39]. Another study also showed that impaired insulin/IGF-1 signaling extends the C. elegans lifespan by promoting mitochondrial proline catabolism, while proline supplementation extends the lifespan [40]. These results suggest that proline directly affects longevity in C. elegans, but change in amino acid metabolism caused by the intracellular proline might have some effect on longevity in yeast. However, little is known about the roles of proline regulating the replicative lifespan of yeast cell. We previously found that deletion of genes involved in converting glutamate to γ-aminobutyric acid could increase the replicative lifespan [31] and lead to increased conversion of glutamate to α-ketoglutarate and other TCA cycle intermediates that maintain mitochondrial respiratory function. The replicative lifespan is also regulated by genes encoding metabolic enzymes, the hexokinase isoenzyme 2 Hxk2p, the alcohol dehydrogenase Adh1p, and the saccharopine dehydrogenase Lys9p [27][28][29]. Yeast is a unique and powerful model for investigation of the effects of these genes on lifespan in eukaryotic cells. Furthermore, breeding of industrial yeast strains with accumulation of proline, which is correlated with lifespan extension, could contribute to an improvement in fermentation ability and compound productivity.
MATERIALS AND METHODS
Strains and media
The S. cerevisiae strains used in this study were derived from BY4741 or CAY29, which are S288C-derivative laboratory strains, and are listed in Table S1. The deletion strains were obtained from the Yeast MATa Collection (Open Biosystems, AL) for BY1-put1, BY1-car2, BY1-pro1, and BY1-pro2 and P. O. Ljungdahl for CAY191 [37]. The other mutant and overexpression strains (BY1-PRO1-I150T, BY1-put1-PRO1-I150T, BY1-MSN2OE, BY1-MSN2OE-put1, and BY1-msn2/4) were constructed in this study (Table S1). Gene manipulations such as gene overexpression, and gene deletions were carried out through genomic integration as described previously [41]. Oligonucleotide primers used in this study are listed in Table S2. We first integrated a mutated PRO1 gene by homologous recombination by the URA3 marker. In order to construct PRO1-I150T mutant strains, a linearized plasmid, pRS406-I150TPRO1 [42], which expresses the Ile150Thr variant of Pro1p, was introduced into strain BY4741 or BY1-put1. After the Ura+ transformants grown on SC-uracil medium (20 g/L glucose, 6.7 g/L Bacto yeast nitrogen base without amino acids [Difco Laboratories, Detroit, MI], 2 g/L drop-out mixture) were cultured in YPD medium (20 g/L glucose, 10 g/L Bacto yeast extract, 20 g/L Bacto peptone) at 30°C for 24 h with shaking, diluted to the same media, incubated for several days, strains BY1-PRO1-I150T and BY1-put1-PRO1-I150T that have excised the plasmid and lost one of the two copies of the duplicated region by homologous crossover were obtained from 5-fluoroorotic acid-containing plates [43].
–
The MSN2 overexpression cassette consisting of the URA3 marker (containing both native promoter and terminator of the URA3 gene) and a constitutive, a strong promoter of the TDH3 gene (PTDH3) was amplified via polymerase chain reaction (PCR) using a set of primers (MSN2_up_F and MSN2_from_1_to_534_R). This cassette was then integrated between the native promoter of MSN2 gene (PMSN2) and the open reading frame of MSN2 gene through yeast transformation. Following that, positive clones were confirmed using another set of primers (TDH3_up_F(-132_-108) and MSN2_R2). For MSN2 deletion, this pair of primers was used (MSN2+URA3-Fw and MSN2+URA3-Rv) to amplify the disruption cassette. For gene deletions, plasmid pUG6 harboring geneticin-resistant gene was used as a template to amplify the deletion cassettes using specific sets of primers (Fw Δmsn4-kanMX and Rv Δmsn4-kanMX). YPD medium was used for routine cultures, and G418 (200 µg/mL) was added to YPD medium when required.
–
Replicative lifespan determination
The replicative lifespan was assayed as previously described [38]. A sample of yeast cells was spread onto YPD agar plates containing 10 μg/mL phloxine B. Virgin daughter cells were selected and subjected as sample cells to lifespan analysis. Daughter cells were removed with a dissecting needle and scored every 2 h. For each of at least 48 cell lines, buds from each mother cell were counted until division of living cells ceased on YPD agar plates or cells were stained with phloxine B. Table 1 shows all of the replicative lifespan data. For statistical analysis, lifespan data sets were compared by a Wilcoxon rank-sum test. Two strains were stated to have a significant difference in lifespan when p < 0.05.
–
–
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
The isolation of total RNA from yeast cells and subsequent RT-qPCR analysis were carried out as previously described [38]. Total RNA was extracted from yeast cells using a RNeasy Mini Kit (Qiagen, Valencia, CA, USA). Extracted total RNAs were subjected to cDNA synthesis using a PrimeScript RT Master Mix (Takara Bio, Shiga, Japan) with random primers. qPCR was carried out using a SYBR Premix Ex TaqII (Takara Bio) on a Thermal Cycler Dice Real Time System II (Takara Bio). The relative mRNA concentration was normalized to that of the RDN18 gene transcript as an internal control. For statistical analysis, RT-qPCR data sets were compared by a Student's t-test. Two strains were stated to have a significant difference in lifespan when p < 0.05.
–
Amino acid quantification
For the determination of intracellular proline and other amino acids, yeast cells were grown in 5 mL YPD medium at 30°C to the log and stationary phases for 8 h and 24 h, respectively, with shaking (120 rpm); 500 μL cell suspension were removed, the cells were washed twice with 0.9% NaCl and suspended in 0.5 mL distilled water. The 1.5 mL microcentrifuge tube containing cells was transferred to a boiling water bath, and intracellular amino acids were extracted by boiling for 20 min. After centrifugation (15,000 x g, 5 min, 4°C), each supernatant was analyzed quantitatively using an amino acid analyzer (JLC500/V, Jeol, Tokyo, Japan). Amino acid contents were calculated as a percentage of dry weight. For statistical analysis, quantification data sets were compared by a Dunnett's test. Two strains (each of mutants vs. the wild-type strain) were stated to have a significant difference in amino acid contents when p < 0.05.
–
Multivariate Analysis
The data sets from the amino acid analysis were judged in all cases by principal component analysis (PCA) and orthogonal projection to latent structure (OPLS) using SIMCA-P+ 12.0.1 (Umetrics, Sweden).
REFERENCES
- Poolman B and Glaasker E (1998). Regulation of compatible solute accumulation in bacteria. Mol Microbiol 29(2): 397-407. doi: 10.1046/j.1365-2958.1998.00875.x
- Csonka LN (1989). Physical and genetic responses of bacteria to osmotic stress. Microbiol Rev 53(1): 121–147. PMID: 2651863
- Ignatova Z and Gierasch LM (2006). Inhibition of protein aggregation in vitro and in vivo by a natural osmoprotectant. Proc Natl Acad Sci USA 103(36): 13357-13361. doi: 10.1073/pnas.0603772103
- Kaul S, Sharma SS and Mehta IK (2008). Free radical scavenging potential of L-proline: evidence from in vitro assays. Amino Acids 34(2): 315–320. doi: 10.1007/s00726-006-0407-x
- Csonka LN and Hanson AD (1991). Prokaryotic osmoregulation: genetics and physiology. Annu Rev Microbiol 45: 569–606. doi: 10.1146/annurev.mi.45.100191.003033
- Tomenchok DM and Brandriss MC (1987). Gene-enzyme relationships in the proline biosynthetic pathway of Saccharomyces cerevisiae. J Bacteriol 169(12): 5364–5372. doi: 10.1128/jb.169.12.5364-5372.1987
- Brandriss MC and Falvey DA (1992). Proline biosynthesis in Saccharomyces cerevisiae: analysis of the PRO3 gene, which encodes 1-pyrroline-5-carboxylate reductase. J Bacteriol 174(11): 3782-3788. doi: 10.1128/jb.174.15.5176b.1992
- Degols G, Jauniaux JC and Wiame JM (1987). Molecular characterization of transposable-element-associated mutations that lead to constitutive L-ornithine aminotransferase expression in Saccharomyces cerevisiae. Eur J Biochem 165(2): 289-296. doi: 10.1111/j.1432-1033.1987.tb11440.x
- Takagi H (2008). Proline as a stress protectant in yeast: physiological functions, metabolic regulations, and biotechnological applications. Appl Microbiol Biotechnol 81(2): 211–223. doi: 10.1007/s00253-008-1698-5
- Kaino T and Takagi H (2008). Gene expression profiles and intracellular contents of stress protectants in Saccharomyces cerevisiae under ethanol and sorbitol stresses. Appl Microbiol Biotech 79(2): 273-283. doi: 10.1007/s00253-008-1431-4
- Morita Y, Nakamori S and Takagi H (2003). L-Proline accumulation and freeze tolerance of Saccharomyces cerevisiae are caused by a mutation in the PRO1 gene encoding γ-glutamyl kinase. Appl Environ Microbiol 69(1): 212–219. doi: 10.1128/AEM.69.1.212-219.2003
- Sekine T, Kawaguchi A, Hamano Y and Takagi H (2007). Desensitization of feedback inhibition of the Saccharomyces cerevisiae γ-glutamyl kinase enhances proline accumulation and freezing tolerance. Appl Environ Microbiol 73(12): 4011-4019. doi: 10.1128/AEM.00730-07
- Takagi H, Sakai K, Morida K and Nakamori S (2000). Proline accumulation by mutation or disruption of the proline oxidase gene improves resistance to freezing and desiccation stresses in Saccharomyces cerevisiae. FEMS Microbiol Lett 184(1): 103-108. doi: 10.1111/j.1574-6968.2000.tb08998.x
- Takagi H, Iwamoto F and Nakamori S (1997). Isolation of freeze tolerant laboratory strains of Saccharomyces cerevisiae from proline analogue-resistant mutants. Appl Microbiol Biotechnol 47(4): 405-411. doi: 10.1007/s002530050948
- Terao Y, Nakamori S and Takagi H (2003). Gene dosage effect of L-proline biosynthetic enzymes on L-proline accumulation and freeze tolerance in Saccharomyces cerevisiae. Appl Environ Microbiol 69(11): 6527-6532. doi: 10.1128/AEM.69.11.6527-6532.2003
- Takagi H, Takaoka M, Kawaguchi A and Kubo Y (2005). Effect of L-proline on sake brewing and ethanol stress in Saccharomyces cerevisiae. Appl Environ Microbiol 7 (12): 8656-8662. doi: 10.1128/AEM.71.12.8656-8662.2005
- Sasano Y, Haitani Y, Ohtsu I, Shima J and Takagi H (2012). Proline accumulation in baker's yeast enhances high-sucrose stress tolerance and fermentation ability in sweet dough. Int J Food Microbiol 152(1-2): 40-43. doi: 10.1016/j.ijfoodmicro.2011.10.004
- Takagi H, Taguchi J and Kaino T (2016). Proline accumulation protects Saccharomyces cerevisiae cells in the stationary phase from ethanol stress by reducing reactive oxygen species levels. Yeast 33(8): 355-363. doi: 10.1002/yea.3154
- Tsolmonbaatar A, Hashida K, Sugimoto Y, Watanabe D, Furukawa S and Takagi H (2016). Isolation of baker's yeast mutants with proline accumulation that showed enhanced tolerance to baking-associated stresses. Int J Food Microbiol 238: 233-240. doi: 10.1016/j.ijfoodmicro.2016.09.015
- Chen YF, Wu CY, Kao CH and Tsai TF (2010). Longevity and lifespan control in mammals: lessons from the mouse. Ageing Res Rev 9(suppl 1): S28-35. doi: 10.1016/j.arr.2010.07.003
- He Y and Jasper H (2014). Studying aging in Drosophila. Methods 68(1): 129-133. doi: 10.1016/j.ymeth.2014.04.008
- Tissenbaum HA (2015). Using C. elegans for aging research. Invertebr Reprod 59(sup1): 59-63. doi: 10.1080/07924259.2014.940470
- He C, Zhou C and Kennedy BK (2018). The yeast replicative aging model. Biochim Biophys Acta Mol Basis Dis 1864(9 Pt A): 2690-2696. doi: 10.1016/j.bbadis.2018.02.023
- Mortimer RK and Johnston JR (1959). Life span of individual yeast cells. Nature 183(4677): 1751-1752. doi: 10.2172/4329664
- Longo VD, Shadel GS, Kaeberlein M and Kennedy BK (2012). Replicative and chronological aging in Saccharomyces cerevisiae. Cell Metab 16(1): 18-31. doi: 10.1016/j.cmet.2012.06.002
- Mirisola MG, Braun RJ and Petranovic D (2014). Approaches to study yeast cell aging and death. FEMS Yeast Res 14(1): 109-118. doi: 10.1111/1567-1364.12112
- Lin SJ, Defossez PA and Guarente L (2000). Requirement of NAD and SIR2 for life-span extension by calorie restriction in Saccharomyces cerevisiae. Science 289(5487): 2126-2128. doi: 10.1126/science.289.5487.2126
- Kaeberlein M (2005). Regulation of yeast replicative life span by TOR and Sch9 in response to nutrients. Science 310(5751): 1193-1196. doi: 10.1126/science.1115535
- Smith ED, Tsuchiya M, Fox LA, Dang N, Hu D, Kerr EO, Johnston ED, Tchao BN, Pak DN, Welton KL, Promislow DEL, Thomas JH, Kaeberlein M and Kennedy BK (2008). Quantitative evidence for conserved longevity pathways between divergent eukaryotic species. Genome Res 18(4): 564-570. doi: 10.1101/gr.074724.107
- Yoshida R, Tamura T, Takaoka C, Harada K, Kobayashi A, Mukai M and Fukusaki E (2010). Metabolomics-based systematic prediction of yeast lifespan and its application for semi-rational screening of ageing-related mutants. Aging Cell 9(4): 616-625. doi: 10.1111/j.1474-9726.2010.00590.x
- Kamei Y, Tamura T, Yoshida R, Ohta S, Fukusaki E and Mukai Y (2011). GABA metabolism pathway genes, UGA1 and GAD1, regulate replicative lifespan in Saccharomyces cerevisiae. Biochem Biophys Res Commun 407(1): 185-190. doi: 10.1016/j.bbrc.2011.02.136
- Tai A, Kamei Y and Mukai Y (2017). The forkhead-like transcription factor (Fhl1p) maintains yeast replicative lifespan by regulating ribonucleotide reductase 1 (RNR1) gene transcription. Biochem Biophys Res Commun 488(1): 218-223. doi: 10.1016/j.bbrc.2017.05.038
- Fabrizio P and Longo VD (2007). The chronological life span of Saccharomyces cerevisiae. Meth Mol Biol 371: 89-95. doi: 10.1007/978-1-59745-361-5_8
- Urban J, Soulard A, Huber A, Lippman S, Mukhopadhyay D, Deloche O, Wanke V, Anrather D, Ammerer G, Riezman H, Broach JR, De Virgilio C, Hall MN and Loewith R (2007). Sch9 is a major target of TORC1 in Saccharomyces cerevisiae. Mol Cell 26(5): 663-674. doi: 10.1016/j.molcel.2007.04.020
- Schmitt AP and McEntee K (1996). Msn2p, a zinc finger DNA-binding protein, is the transcriptional activator of the multistress response in Saccharomyces cerevisiae. Proc Natl Acad Sci USA 93(12): 5777-5782. doi: 10.1073/pnas.93.12.5777
- Martínez-Pastor MT, Marchler G, Schüller C, Marchler-Bauer A, Ruis H and Estruch F (1996). The Saccharomyces cerevisiae zinc finger proteins Msn2p and Msn4p are required for transcriptional induction through the stress response element (STRE). EMBO J 15(9): 2227-2235. doi: 10.1002/j.1460-2075.1996.tb00576.x
- Andréasson C, Neve EP and Ljungdahl PO (2004). Four permeases import proline and the toxic proline analogue azetidine-2-carboxylate into yeast. Yeast 21(3): 193-199. doi: 10.1002/yea.1052
- Kamei Y, Tamada Y, Nakayama Y, Fukusaki E and Mukai Y (2014). Changes in transcription and metabolism during the early stage of replicative cellular senescence in budding yeast. J Biol Chem 289(46): 32081-32093. doi: 10.1074/jbc.M114.600528
- Edwards C, Canfield J, Copes N, Brito A, Rehan M, Lipps D, Brunquell J, Westerheide SD and Bradshaw PC (2015). Mechanisms of amino acid-mediated lifespan extension in Caenorhabditis elegans. BMC Genet 16(8): doi: 10.1186/s12863-015-0167-2.
- Zarse K, Schmeisser S, Groth M, Priebe S, Beuster G, Kuhlow D, Guthke R, Platzer M, Kahn CR and Ristow M (2012). Impaired insulin/IGF1 signaling extends life span by promoting mitochondrial L-proline catabolism to induce a transient ROS signal. Cell Metab 15(4): 451-465. doi: 10.1016/j.cmet.2012.02.013
- Janke C, Magiera MM, Rathfelder N, Taxis C, Reber S, Maekawa H, Moreno-Borchart A, Doenges G, Schwob E, Schiebel E and Knop M (2004). A versatile toolbox for PCR-based tagging of yeast genes: New fluorescent proteins, more markers and promoter substitution cassettes. Yeast 21(11): 947-962. doi: 10.1002/yea.1142
- Kaino T, Tateiwa T, Mizukami-Murata S, Shima J and Takagi H (2008). Self-cloning baker's yeasts that accumulate proline enhance freeze tolerance in doughs. Appl Environ Microbiol 74(18): 5845-5849. doi: 10.1128/AEM.00998-08
- Sikorski RS and Boeke JD (1991). In vitro mutagenesis and plasmid shuffling: from cloned gene to mutant yeast. Methods Enzymol 194: 302-318. doi: 10.1016/0076-6879(91)94023-6
- Nishida I, Watanabe D and Takagi H (2016). Putative mitochondrial α-ketoglutarate-dependent dioxygenase Fmp12 controls utilization of proline as an energy source in Saccharomyces cerevisiae. Microb Cell 3(10): 522-528. doi: 10.15698/mic2016.10.535
- Andréasson C and Ljungdahl PO (2002). Receptor-mediated endoproteolytic activation of two transcription factors in yeast. Genes Dev 16(24): 3158-3172. doi: 10.1101/gad.239202
SUPPLEMENTAL INFORMATION
![]() Download Supplemental Information
Download Supplemental Information
ACKNOWLEDGMENTS
We are grateful to P. O. Ljungdahl (Stockholm University) for the generous gift of yeast strains. This work was supported by a Grant-in-Aid for Challenging Exploratory Research (19K22282) from Ja-pan Society for the Promotion of Science (JSPS) to HT and a grant from the Project of the NARO Bio-oriented Technology Research Advancement Institution (Re-search program on development of innovative technol-ogy) (30017B) to HT.
COPYRIGHT
© 2019

Proline metabolism regulates replicative lifespan in the yeast Saccharomyces cerevisiae by Mukai et al. is licensed under a Creative Commons Attribution 4.0 International License.