News and thoughts:
Microbial Cell, Vol. 7, No. 3, pp. 62 - 65; doi: 10.15698/mic2020.03.709
Raman-based sorting of microbial cells to link functions to their genes
1 Institute for Environmental Engineering, Department of Civil, Environmental and Geomatic Engineering, ETH Zurich, Zurich, Switzerland.
2 University of Vienna, Centre for Microbiology and Environmental Systems Science, Department of Microbiology and Ecosystem Science, Althanstrasse 14, 1090 Vienna, Austria.
3 Center for Microbial Communities, Department of Chemistry and Bioscience, Aalborg University, 9220 Aalborg, Denmark.
Keywords: single cell Raman microspectroscopy, stable isotope probing, optical tweezers, microfluidic sorting, metagenomics, optofluidics.
Abbreviations:
CARS – coherent anti-Stokes Raman spectroscopy;
D – deuterium;
DMSP – dimethylsulfoniopropionate;
PDMS – polydimethylsiloxane;
RACS – Raman-activated cell sorting;
SIP – stable isotope probing;
WGA – whole genome amplification.
Received originally: 18/11/2019 Received in revised form: 24/01/2020
Accepted: 29/01/2020
Published: 10/02/2020
Correspondence:
Roman Stocker, Institute for Environmental Engineering, Department of Civil, Environmental and Geomatic Engineering, ETH Zurich, Zurich, Switzerland; romanstocker@ethz.ch
Conflict of interest statement: The authors declare no conflict of interest.
Please cite this article as: Kang Soo Lee, Michael Wagner and Roman Stocker (2020). Raman-based sorting of microbial cells to link functions to their genes. Microbial Cell 7(3): 62-65. doi: 10.15698/mic2020.03.709
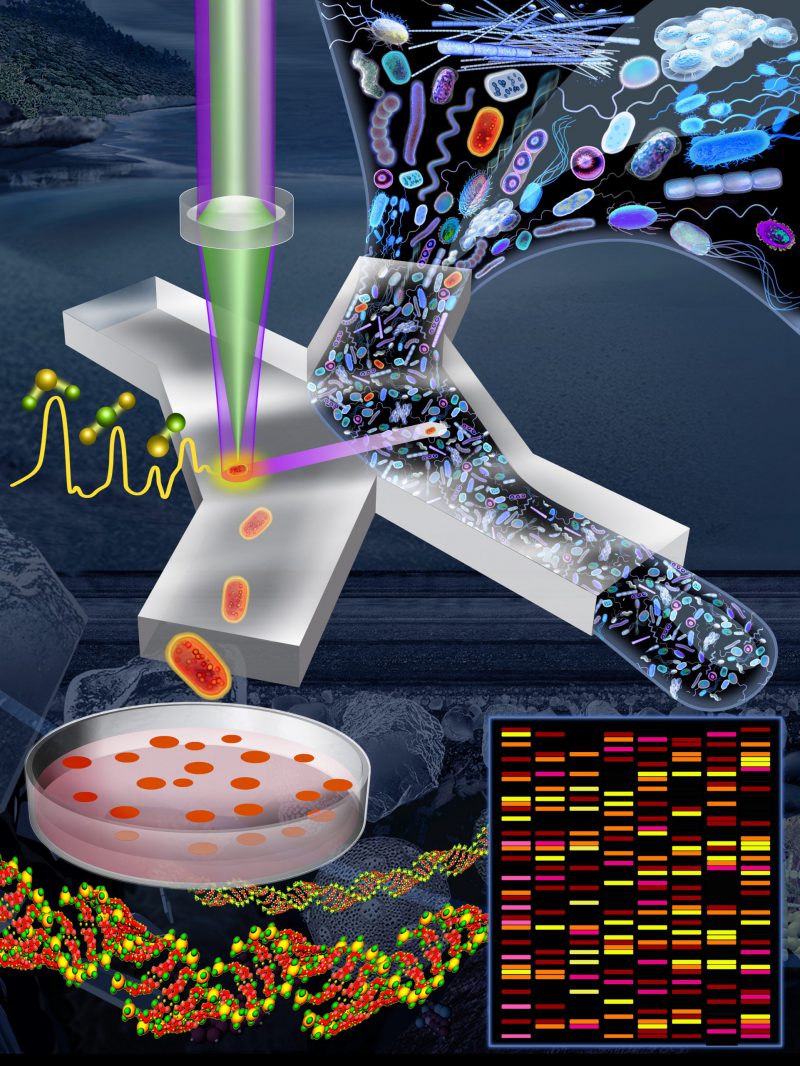
In our recent work, we developed an optofluidic platform that allows a direct link to be made between the phenotypes (functions) and the genotypes (genes) of microbial cells within natural communities. By combining stable isotope probing, optical tweezers, Raman microspectroscopy, and microfluidics, the platform performs automated Raman-based sorting of taxa from within a complex community in terms of their functional properties. In comparison with manual sorting approaches, our method provides high throughput (up to 500 cells per hour) and very high sorting accuracy (98.3 ± 1.7%), and significantly reduces the human labour required. The system provides an efficient manner to untangle the contributions of individual members within environmental and host-associated microbiomes. In this News and Thoughts, we provide an overview of our platform, describe potential applications, suggest ways in which the system could be improved, and discuss future directions in which Raman-based analysis of microbial populations might be developed.
There have been major recent technical advances toward understanding the composition of natural microbial communities and the contributions of individual organisms to their functioning. Stable isotope probing (SIP) provides a means to identify the organisms responsible for performing key functions within complex communities, and can provide information at the resolution of a single cell. Targeted sorting of cells from natural communities in terms of their functional properties and the use of the sorted cells for downstream DNA analysis or cultivation is becoming feasible, and makes it possible to establish a direct link between the phenotype (function) and the genotype (genome) of the members of a community. However, the approach is still in its infancy and major issues have precluded broader adoption, the most critical ones being the low throughput [1][2] and the loss of cell viability induced during phenotypic identification [2][3].
–
In our recent publication [4], we introduced a Raman-activated cell sorting (RACS) pipeline that enables microbial ecologists to link microbial metabolic phenotypes to their genotypes in a high-throughput and non-destructive manner (Figure 1). Within a complex natural microbial community, cells that are able to metabolize a particular compound can be labelled with deuterium (D) by incubating a community sample in a D2O-containing ‘minimal' (i.e., lacking nutrients) medium supplemented with the unlabelled compound of interest (e.g., glucose, mucin). After a short incubation, only active cells that were able to metabolize the compound of interest become D-labelled, resulting in the appearance of a signature peak in their Raman spectra (the carbon–deuterium ‘C–D' peak; the spectral region of 2,040–2,300 cm-1), which is easily detected using Raman microspectroscopy. By coupling SIP technology with an automated Raman-based sorting platform that uses optical tweezers and microfluidics to manipulate cells, we demonstrated that it is possible to achieve high-throughput sorting (200–500 cells/h; two orders of magnitude greater than by manual sorting) and the collection of D-labelled cells with high sorting accuracy (98.3 ± 1.7%). Our platform is suitable to sort individual cells for genomic analyses such as 16S rRNA gene sequencing and metagenomics, but also, since live cells are obtained, for cultivation and further ecological characterization or selection. We characterized and optimized the platform using laboratory strains of four model species (two intestinal, one soil and one marine – Escherichia coli, Salmonella typhimurium, Bacillus subtilis, and Marinobacter adhaerens). We then demonstrated the complete pipeline – from sample preparation to DNA analysis – to sort and identify mucin degraders within a complex community sample from the mouse colon. Our approach revealed that organisms belonging to the family Muribaculaceae, an abundant but under-characterized family present in the guts of homoeothermic animals, are key players in mucin degradation. Finally, we demonstrated the versatility of the system by sorting cells from a marine sample in terms of their cytochrome c signal. The few other cell sorting methods in the literature have applications limited to pure cultures (i.e., cells that can be cultured in the laboratory), large-sized cells (e.g., eukaryotes), and/or cells that contain a specific biochemical compound that generates a strong signal from ‘resonance' Raman scattering (e.g., carotenoids) [2][5][6][7][8][9]. To the best of our knowledge, our platform is the first that works for diverse molecular signals in natural samples composed of microbial cells.
–
–
A key parameter for successful cultivation of sorted cells is the laser power chosen for the Raman measurements and optical tweezers. However, recommendation of a universal laser power suitable for any application is difficult, because the magnitude of photophoretic damage depends both on the bacterial type [10], and on the wavelength used [11]. We used 532-nm (green) and 1,064-nm (infrared) lasers for Raman measurement and optical tweezing, respectively. For the Raman measurement, we employed a 15-mW laser (which yields 65.5 MW/m2 when using a 60× 1.2 NA objective; the photophoretic damage is dependent on the laser power density, not the power itself). This value was chosen by gradually changing the laser power to obtain the maximum power that did not cause lysis of an optically trapped cell over 10 s (sorting itself took a maximum of 7–8 s for each cell). Similarly, 400 mW was chosen for the 1,064-nm laser. Culture of sorted cells showed that they suffered no loss of viability. Empirical trials of this kind should be used to choose the power to employ for a given application.
–
The platform has many potential applications. For example, in host-associated microbiome research it provides a means to disentangle the symbiotic interaction between two partners (e.g., algae and associated bacteria). The host can be D-labelled by pre-culturing in D2O-containing medium, followed by a co-culture with the partner in non-D2O medium. Sorting the microbial partners in terms of D-labelling and subsequent DNA analysis can elucidate which taxa consume biomolecules from the host, and vice versa when the pre-culturing for D-labelling begins with the partner. In addition to classical deuterium probing, detection of those bacteria able to degrade particular compounds is possible by using labelled versions of those compounds: 13C-, 15N-, and D-labelled compounds can all be detected using Raman microspectroscopy. RACS thus provides a powerful means to determine the participants and the currency of symbiotic exchange. The same approach offers an efficient method to determine the bacteria performing important ecological or industrial functions, for example, using labelled dimethylsulfoniopropionate (DMSP) to detect DMSP-consuming bacteria within samples from the ocean, or labelled phosphate compounds such as Na2DPO4 to identify polyphosphate-accumulating organisms (PAO) in samples from wastewater treatment plants [12]. Furthermore, our sorting platform is extremely versatile: the spectral region in the sorting software can easily be tuned to use the presence of other functional molecules as the sorting criterion. Any molecule with a characteristic Raman spectrum is a potential target, for example, cytochrome c peaks (750; 1,127; 1,314; and 1,585 cm-1) as in our paper; calcium dipicolinate peaks (1,016; 1,400; 1,447; and 1,568 cm-1) to distinguish bacterial endospores from vegetative cells [13]; or the peak of surface-enhanced Raman scattering (SERS) nanotags used to label targeted pathogenic microbes [14].
–
Our platform can potentially be used to sort diverse microbes including algae (e.g., diatoms, cyanobacteria), fungi (e.g., yeast), and even mammalian cells (e.g., blood cells, tumor cells) that are larger than the bacterial cells used in our paper [4]. Raman microspectroscopy is based on a point measurement (i.e., the signal comes from the laser spot focused using an objective), and thus in general, the larger the cell (occupying more space in the measurement point), the greater the Raman signal intensity. The other crucial parameter is the optical tweezing efficiency (which determines the ability to hold the cell for the measurement period — key to reliable and reproducible Raman measurement). In general, the larger the cell, the greater the optical tweezing efficiency. Therefore, sorting of larger cells in our platform would be feasible. By the same considerations, there may be a lower limit on the applicable cell size when the optical tweezing force is not sufficient to capture the cell for the Raman measurement. We have successfully used the platform on cells down to a size of ~1 µ#x03BC;m, but the use of smaller microbes, such as viruses, may not be possible. Heterogeneity in cell morphology (e.g., spherical, rod-shaped, filamentous) is of lesser influence, because a cell of arbitrary shape is in any case trapped at the equilibrium position within the optical gradient force field [15][16].
–
Improvements to the experimental setup could enhance its suitability to sort anaerobic samples, such as the mouse colon community in our work. For such samples, it is important that the platform maintains anaerobic conditions. However, in our experiment, even after being exposed to aerobic conditions in the gas-permeable polydimethylsiloxane (PDMS) microfluidic device and polyethylene (PE) tubing, the collected mouse colon taxa were still viable after sorting (unpublished data). This might be attributed to the brevity of the exposure (~1 h) to aerobic conditions, another advantage of rapid sorting. Nevertheless, the ability to maintain anaerobic conditions could be further improved by making the remaining components less gas permeable, such as by using polyetheretherketone (PEEK) or ethylene tetrafluoroethylene (ETFE) tubing, and the coating of the inner surface of the PDMS channels with gas-impermeable material (e.g., parylene C [17]) or by enclosing the whole device with acrylic, as in the anaerobic intestine-on-a-chip [18].
–
Future development of the platform could foster even wider applicability. A key feature in many applications is throughput, which is strongly related to the Raman signal sensitivity of the platform. For metagenomics, the collected cells after sorting are lysed and their DNA amplified by whole genome amplification (WGA). If the throughput were to be improved by 1–2 orders of magnitude (to a rate of a few tens of thousands of cells per hour), DNA amplification would no longer be necessary. Additional valuable information would thus be made available, such as the relative abundance of the taxa within a community; information that cannot be obtained after WGA. There is room for the necessary technical improvements. Methods employing ‘coherent' Raman scattering microspectroscopy, such as stimulated Raman spectroscopy (SRS), femtosecond stimulated Raman spectroscopy (FSRS), and coherent anti-Stokes Raman spectroscopy (CARS), provide powerful tools to increase the intrinsically weak spontaneous Raman scattering by more than thousand-fold. Fourier transform (FT)-CARS was recently used to detect astaxanthin productivity and photosynthetic dynamics of green algae in a microfluidic device that forms a single-file cell stream using a surface acoustic wave [19]. By avoiding the need to capture each cell for measurement, the device achieved a very high recording speed of cells as they passed in this flow (~2,000 events per second), and this technology would be applicable for Raman-activated sorting if two technical challenges were to be addressed. First, only large algal cells (Haematococcus lacustris and Euglena gracilis) were measured. There are many three-dimensional focusing techniques in microfluidics (e.g., hydrodynamic, acoustic, electric, and geometry-induced); however, it is still not an easy matter to tightly focus smaller bacterial cells (~1 µ#x03BC;m) in a narrow region of the flow. As the Raman signal is strongly dependent upon the position of the cell as it passes through the laser detection spot, the platform must be engineered to make tight focusing possible. Second, the existing system is a cell counter, and further engineering would be necessary to enable the platform to sort cells after the CARS measurement without greatly sacrificing throughput. In addition to these developments in the hardware, the software could also be made more versatile in the choice of the sorting criteria, by implementing machine-learning algorithms. In some situations, such as for cytochrome c–containing cells in marine community samples, cells show an inconsistent background in their Raman spectra depending on conditions (e.g., oxidation state), and thus universal sorting criteria are difficult to define by the user. In this instance, the software platform could first measure the spectra of an initial “learning” population of cells. It could then determine the sorting criterion by categorizing those data into a few sub-populations (one of which is the target), and then perform the actual sorting based upon this initial classification.
–
We anticipate that integration of our platform with recent technological advances in spectral data processing (e.g., machine learning) and electronics and photonics (e.g., coherent Raman system) will further improve its accuracy and throughput, opening new avenues for research in ecological, medical, and industrial microbiology. Already, armed with our RACS platform, researchers have a new efficient tool to understand the functioning of microbial communities.
REFERENCES
- Berry D, Mader E, Lee TK, Woebken D, Wang Y, Zhu D, Palatinszky M, Schintlmeister A, Schmid MC, Hanson BT, Shterzer N, Mizrahi I, Rauch I, Decker T, Bocklitz T, Popp J, Gibson CM, Fowler PW, Huang WE, and Wagner M (2015). Tracking heavy water (D2O) incorporation for identifying and sorting active microbial cells. Proc Natl Acad Sci USA 112(2): E194-203. doi: 10.1073/pnas.1420406112
- Jing X, Gou H, Gong Y, Su X, Xu L, Ji Y, Song Y, Thompson IP, Xu J, and Huang WE (2018). Raman-activated cell sorting and metagenomic sequencing revealing carbon-fixing bacteria in the ocean. Environ Microbiol 20(6):2241-2255. doi: 10.1111/1462-2920
- Hatzenpichler R, Connon SA, Goudeau D, Malmstrom RR, Woyke T, and Orphan VJ (2016). Visualizing in situ translational activity for identifying and sorting slow-growing archaeal-bacterial consortia. Proc Natl Acad Sci USA 113(28): E4069-E4078. doi: 10.1073/pnas.1603757113
- Lee KS, Palatinszky M, Pereira FC, Nguyen J, Fernandez VI, Mueller AJ, Menolascina F, Daims H, Berry D, Wagner M, and Stocker R (2019). An automated Raman-based platform for the sorting of live cells by functional properties. Nat Microbiol 4(6): 1035-1048. doi: 10.1038/s41564-019-0394-9
- Wang Y, Ji Y, Wharfe ES, Meadows RS, March P, Goodacre R, Xu J, and Huang WE (2013). Raman activated cell ejection for isolation of single cells. Anal Chem 85(22): 10697-10701. doi: 10.1021/ac403107p
- Wang X, Ren L, Su Y, Ji Y, Liu Y, Li C, Li X, Zhang Y, Wang W, Hu Q, Han D, Xu J, and Ma B (2017). Raman-activated droplet sorting (RADS) for label-free high-throughput screening of microalgal single-cells. Anal Chem 89(22): 12569-12577. doi: 10.1021/acs.analchem.7b03884
- Zhang P, Ren L, Zhang X, Shan Y, Wang Y, Ji Y, Yin H, Huang WE, Xu J, and Ma B (2015). Raman-activated cell sorting based on dielectrophoretic single-cell trap and release. Anal Chem 87(4): 2282-2289. doi: 10.1021/ac503974e
- McIlvenna D, Huang WE, Davison P, Glidle A, Cooper J, and Yin H (2016). Continuous cell sorting in a flow based on single cell resonance Raman spectra. Lab Chip 16(8): 1420-1429. doi: 10.1039/c6lc00251j
- Wang Y, Ji Y, Wharfe ES, Meadows RS, March P, Goodacre R, Xu J, and Huang WE (2013). Raman activated cell ejection for isolation of single cells. Anal Chem 85(22) 10697-10701. doi: 10.1021/ac403107p
- Yuan X, Song Y, Song Y, Xu J, Wu Y, Glidle A, Cusack M, Ijaz UZ, Cooper JM, Huang WE, and Yin H (2018). Effect of laser irradiation on cell function and its implications in Raman spectroscopy. Appl Environ Microbiol 84(8) e02508-17. doi: 10.1128/AEM.02508-17
- Dholakia K and Reece P (2006). Optical micromanipulation takes hold. Nano Today 1(1) 18–27. doi: 10.1016/S1748-0132(06)70019-6
- Fernando EY, McIlroy SJ, Nierychlo M, Herbst FA, Petriglieri F, Schmid MC, Wagner M, Nielsen JL, and Nielsen PH (2019). Resolving the individual contribution of key microbial populations to enhanced biological phosphorus removal with Raman-FISH. ISME J 13(8):1933-1946. doi: 10.1038/s41396-019-0399-7
- Stöckel S, Meisel S, Böhme R, Elschner M, Rösch P, and Popp J (2009). Effect of supplementary manganese on the sporulation of Bacillus endospores analysed by Raman spectroscopy. J Raman Spectrosc 40(11): 1469-1477. doi: 10.1002/jrs.2292
- Lin HY, Huang CH, Hsieh WH, Liu LH, Lin YC, Chu CC, Wang ST, Kuo IT, Chau LK, and Yang CY (2014). On-line SERS detection of single bacterium using novel SERS nanoprobes and a microfluidic dielectrophoresis device. Small 10(22): 4700-4710. doi: 10.1002/smll.201401526
- Ling L, Zhou F, Huang L, and Li ZY (2010). Optical forces on arbitrary shaped particles in optical tweezers. J Appl Phys 108(7): 073110. doi: 10.1063/1.3484045
- Landenberger B, Höfemann H, Wadle S, and Rohrbach A (2012). Microfluidic sorting of arbitrary cells with dynamic optical tweezers. Lab Chip 12(17) 3177-3183. doi: 10.1039/C2LC21099A
- Flueckiger J, Bazargan V, Stoeber B, and Cheung KC (2011). Characterization of postfabricated parylene C coatings insde PDMS microdevices. Sensor Actuat B 160(1): 864-874. doi: 10.1016/j.snb.2011.08.073
- Jalili-Firoozinezhad S, Gazzaniga FS, Calamari EL, Camacho DM, Fadel CW, Bein A, Swenor B, Nestor B, Cronce MJ, Tovaglieri A, Levy O, Gregory KE, Breault DT, Cabral JMS, Kasper DL, Novak R, Ingber DE (2019). A complex human gut microbiome cultured in an anaerobic intestine-on-a-chip. Nat Biomed Eng 3(7): 520-531. doi: 10.1038/s41551-019-0397-0
- Hiramatsu K, Ideguchi T, Yonamine Y, Lee S, Luo Y, Hashimoto K, Ito T, Hase M, Park JW, Kasai Y, Sakuma S, Hayakawa T, Arai F, Hoshino Y, and Goda K (2019). High-throughput label-free molecular fingerprinting flow cytometry. Sci Adv 5(1): eaau0241. doi: 10.1126/sciadv.aau0241
ACKNOWLEDGMENTS
We acknowledge support by a US Department of Energy Joint Genome Institute Emerging Technologies Opportunity grant DE-AC02-05CH11231 (to R.S., M.W. and K.S.L.). R.S. acknowledges support from a Gordon and Betty Moore Marine Microbial Initia-tive Investigator Award (#3783) and a grant (#542395) from the Simons Foundation through the Principles of Microbial Ecosystems (PriME) collaborative. M.W. was supported by the European Research Council via the Advanced Grant project NITRICARE 294343. We thank Dr. Russell Naisbit for scientific editing.
COPYRIGHT
© 2020

Raman-based sorting of microbial cells to link functions to their genes by Lee et al. is licensed under a Creative Commons Attribution 4.0 International License.