Research Reports:
Microbial Cell, Vol. 10, No. 8, pp. 170 - 177; doi: 10.15698/mic2023.08.802
Metallothionein Cup1 attenuates nitrosative stress in the yeast Saccharomyces cerevisiae
1 Division of Biological Science, Graduate School of Science and Technology, Nara Institute of Science and Technology, 8916-5 Takayama, Ikoma, Nara 630-0192, Japan.
2 Present address: Department of Biotechnology, Faculty of Bioresource Science, Akita Prefectural University, 241-438 Kaidoubata-Nishi, Shimoshinjo-Nakano, Akita, Akita 010-0195, Japan.
3 Present address: Engineering Biology Research Center, Kobe University, 7-1-48, Minatojima Minami-machi, Chuo-ku, Kobe, Hyogo, 650-0047, Japan.
4 Faculty of Life and Environmental Sciences, Microbiology Research Center for Sustainability, University of Tsukuba, 1-1-1 Tennodai, Tsukuba, Ibaraki 305-8572, Japan.
# The first two authors contributed equally to the manuscript.
Keywords: metallothionein, nitrosative stress, yeast, stress tolerance.
Abbreviations:
MT – Metallothioein,
NO – nitric oxide,
RNS – reactive nitrogen species,
fHb – flavohemoglobin,
GCH2 – GTP cyclohydrolase Ⅱ,
GSNO – S-nitrosoglutathione,
GSNOR – GSNO reductase,
Trx – thioredoxin,
Trr – thioredoxin reductase,
WT – wild-type,
SD – synthetic dextrose,
DAF-FM DA – 4-amino-5-methylamino-2',7'-difluorofluorescein diacetate,
NOC-5 – 3-[2-hydroxy-1-(1-methylethyl)-2-nitrosohydrazino]-1-propanamine,
Grx – glutaredoxin,
Grr – glutaredoxin.
Received originally: 06/04/2023 Received in revised form: 26/06/2023
Accepted: 27/06/2023
Published: 10/07/2023
Correspondence:
Hiroshi Takagi, Division of Biological Science, Graduate School of Science and Technology, Nara Institute of Science and Technology, 8916-5 Takayama, Ikoma, Nara 630-0192, Japan; hiro@bs.naist.jp
Conflict of interest statement:
No potential conflict of interest was reported by the authors.
Please cite this article as: Yuki Yoshikawa, Ryo Nasuno, Naoki Takaya, Hiroshi Takagi (2023). Metallothionein Cup1 attenuates nitrosative stress in the yeast Saccharomyces cerevisiae. Microbial Cell10(8): 170-177. doi: 10.15698/mic2023.08.802
Abstract
Metallothionein (MT), which is a small metal-binding protein with cysteine-rich motifs, functions in the detoxification of heavy metals in a variety of organisms. Even though previous studies suggest that MT is involved in the tolerance mechanisms against nitrosative stress induced by toxic levels of nitric oxide (NO) in mammalian cells, the physiological functions of MT in relation to NO have not been fully understood. In this study, we analyzed the functions of MT in nitrosative stress tolerance in the yeast Saccharomyces cerevisiae. Our phenotypic analyses showed that deletion or overexpression of the MT-encoding gene, CUP1, led to higher sensitivity or tolerance to nitrosative stress in S. cerevisiae cells, respectively. We further examined whether the yeast MT Cup1 in the cell-free lysate scavenges NO. These results showed that the cell-free lysate containing a higher level of Cup1 degraded NO more efficiently. On the other hand, the transcription level of CUP1 was not affected by nitrosative stress treatment. Our findings suggest that the yeast MT Cup1 contributes to nitrosative stress tolerance, possibly as a constitutive rather than an inducible defense mechanism.
INTRODUCTION
Nitric oxide (NO), which is one of the reactive nitrogen species (RNS), functions as a ubiquitous signaling molecule in a variety of biological phenomena [1][2]. The effects of NO on cells can be either beneficial or harmful depending on its concentration. At appropriate levels, NO exerts physiological functions in humans, plants, and microorganisms [3]. Thus, NO induces the relaxation of vascular smooth muscle cells [4], and NO is involved in high temperature stress tolerance in yeast [5][6]. But in excess amounts, NO can induce cellular damage and/or cell death, which is called nitrosative stress. For example, NO induces genetic mutations through DNA strand breakage and/or purine deamination [7]. Due to its high reactivity, NO synthesized by macrophages functions as a weapon to kill pathogenic microbes during infection [8][9]. Therefore, mechanisms to control the concentration of NO or attenuate nitrosative stress are important for the growth and survival of various organisms.
–
A wide variety of NO-degrading and detoxification mechanisms have been reported. NO is oxidized or reduced to NO3– or N2O under aerobic or anaerobic conditions by flavohemoglobin (fHb), respectively [10][11]. Catalase degrades NO to nitrite in the presence of H2O2 even though NO inhibits the activity of catalase via its competitive binding against H2O2 [12]. Recently, we found that catalase was important for nitrosative stress resistance in NADPH-depleted yeast cells [13]. In another recent study, we found that GTP cyclohydrolase II (GCH2), which is the first-step enzyme in the riboflavin biosynthesis pathway, contributes to the nitrosative stress tolerance in yeast through mechanisms in which the reaction product of GCH2, 2,5-diamino-6-(5-phospho-D-ribosylamino)-pyrimidin-4(3H)-one, scavenges RNS [14].
–
NO reacts with thiol-containing compounds and then forms the corresponding S-nitroso molecules. GSH, the reduced form of glutathione, is a thiol-containing peptide that is present in high concentrations in cells, and reacts preferentially with NO to form its S-nitroso derivative, S-nitrosoglutathione (GSNO). GSNO is reductively degraded to GSH and ammonia by GSNO reductase (GSNOR), thioredoxin (Trx), and thioredoxin reductase (Trr) using NADPH as an electron donor [15]. In the filamentous fungus Aspergillus nidulans, an NO-inducible oligopeptide containing many cysteine residues, nitrosothionein, whose transcription is upregulated by RNS, traps NO by its cysteine residues to attenuate nitrosative stress with the aid of Trx, Trr, and NADPH [16]. These results indicate that the intracellular thiol-containing compounds are extremely important for the nitrosative stress tolerance [15].
–
Metallothionein (MT), which has been found in microbes, plants, and mammals, is a small cysteine-rich protein harboring conserved amino acid sequence motifs such as CXC, CXXC, and CCXCC [17][18][19]. MT binds to heavy metals such as zinc, cadmium, and copper via its conserved cysteine-rich motifs to detoxify them [20][21]. MT exists as a complex with transition metal ions like zinc and cadmium and then releases them through reaction with reactive oxygen species or RNS, respectively, which contributes to transition metal homeostasis [22][23][24]. The previous studies reported that overproduction of MT enhanced the RNS resistance of mice cells [25][26]. NO also upregulates the expression level of the MT-encoding gene in rat cells [27]. However, it is still unclear whether the physiological expression level of MT is involved in nitrosative stress tolerance in mammalian cells.
–
In the yeast Saccharomyces cerevisiae, MTs are encoded by the CUP1 or CRS5 gene. S. cerevisiae harbors two copies of the CUP1 gene, CUP1-1 and CUP1-2, of which nucleotide sequences are almost completely identical from 1,533 bp upstream to 269 bp downstream of their coding regions. A previous report suggests that the CUP1 genes are duplicated by segmental duplication in the eighth chromosome [28]. The expression of CUP1 is induced by copper ion via activation of the copper responsive transcriptional activator Ace1 [29]. The Cup1 protein, the gene product of CUP1, functions in the detoxification of metal ions such as copper or cadmium by trapping them [30][31]. CUP1 is also induced by oxidative stress and contributes to oxidative stress tolerance [32]. On the other hand, it was reported that the expression of CRS5 was repressed under oxidative condition [33]. Since NO functions as an oxidant and thus the response mechanisms against nirosative and oxidative stress partly overlap [13][34], Cup1 is likely to be important for nitrosative stress tolerance, even though the physiological roles of Cup1 in yeast under nitrosative stress conditions have not been fully understood.
–
Here, we analyzed the growth phenotypes of S. cerevisiae cells deleting or overexpressing the CUP1 gene under nitrosative stress conditions. In addition, we detected the NO scavenging activity in the Cup1-containing cell-free lysate. Our phenotypic and molecular analyses indicated that Cup1 contributes to the nitrosative stress tolerance by scavenging NO in S. cerevisiae cells.
RESULTS
Yeast metallothionein Cup1 contributes to nitrosative stress tolerance
In order to examine whether the yeast MT Cup1 protects cells from nitrosative stress, we analyzed the nitrosative stress resistance of yeast cells lacking or overexpressing CUP1. We constructed a strain lacking both CUP1-1 and CUP1-2 (cup1Δ strain) (Figure 1A) and a CUP1-1-overexpressing strain. The expression level of the CUP1 gene in each strain was measured by quantitative RT-PCR (RT-qPCR) to confirm the deletion or overexpression of the CUP1 gene (Figure 1B). As a result, the mRNA level of CUP1 was undetectable in the cup1Δ strain, indicating that both the CUP1-1 and CUP1-2 genes were deleted. On the other hand, the CUP1 transcription level was drastically increased in the CUP1-overexpressing strain compared with that in the wild-type (WT) strain, which demonstrates that CUP1 was overexpressed as expected. Subsequently, the growth phenotypes of each strain were analyzed by spot assay (Figure 1C). Deletion or overexpression of the CUP1 gene did not affect the growth of yeast strains on a minimal synthetic dextrose (SD) medium. When yeast strains were cultured with CuSO4, the cup1Δ strain did not grow at all, although the WT strain grew, indicating that CUP1 is involved in the copper resistance, consistent with the previous report [30]. The compensation of copper sensitivity of cup1Δ strain by the overexpression of CUP1his, which encodes Cup1 fused with a hexa-histidine tag at its C-terminus (Cup1-His), demonstrated that Cup1-His normally functions in yeast cells. Next, yeast cells were grown on an acidified medium containing nitrite, under which condition nitrite is converted into RNS [35]. Importantly, the cup1Δ strain exhibited a growth defect compared with WT strain in the presence of RNS, which was recovered by the overexpression of CUP1his, indicating that CUP1 under the control of its physiological expression system is required for nitrosative stress tolerance in yeast. Furthermore, the overexpression of CUP1 improved the growth under nitrosative stress conditions compared with WT strain harboring an empty vector. These results indicated that the CUP1 gene contributed to the nitrosative stress tolerance in a manner dependent on its expression level.
–
–
Cup1 reduces the intracellular NO level by scavenging NO
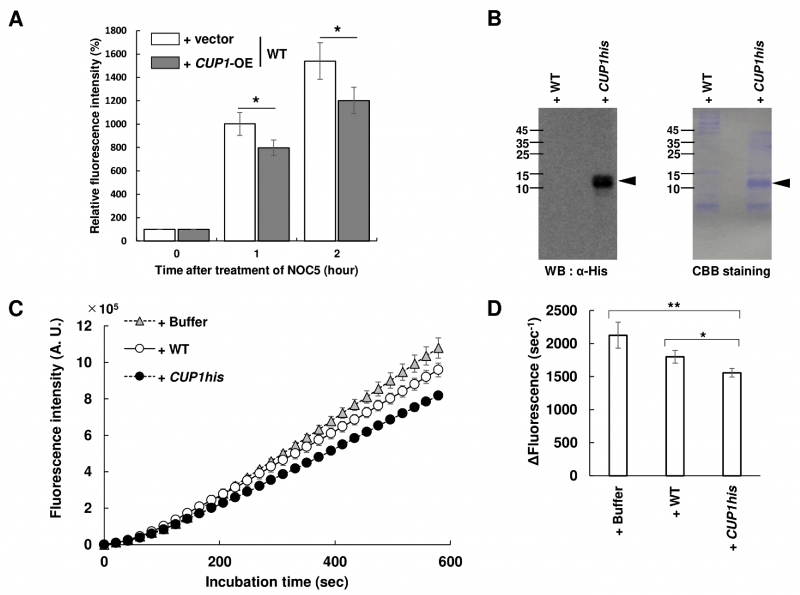
We next analyzed the effect of Cup1 on the intracellular NO content in yeast cells. Yeast cells grown until the exponential growth phase were treated with a cell-permeable fluorescent NO probe, 4-amino-5-methylamino-2',7'-difluorofluorescein diacetate (DAF-FM DA), and then exposed to an NO donor 3-[2-hydroxy-1-(1-methylethyl)-2-nitrosohydrazino]-1-propanamine (NOC-5), followed by flow cytometry (Figure 2A). The results showed that the intracellular fluorescence intensity in WT cells increased over time, which was diminished by the overexpression of CUP1. This suggests that Cup1 decreases the intracellular NO level.
–
We then examined whether the Cup1 protein scavenges NO. For this analysis, we used the WT strain harboring an empty vector and the cup1Δ strain overexpressing CUP1his, which was identical to the strain shown as cup1Δ + CUP1his-OE in Figure 1C (CUP1his strain). Western blotting analysis showed a clear band with the theoretical molecular weight of Cup1-His only in the CUP1his strain, confirming the production of Cup1-His (Figure 2B). SDS-PAGE followed by CBB staining showed a thick band with a molecular weight corresponding to Cup1-His in the CUP1his strain but not in the WT, suggesting the overproduction of Cup1-His protein. Subsequently, a cell-impermeable derivative of DAF-FM DA, DAF-FM, was incubated with NOC-5 in the presence or absence of yeast cell-free lysate and the fluorescence intensity was monitored over time (Figure 2C and 2D). The fluorescence intensity increased slightly more slowly in the presence of lysate from the WT strain than in the presence of the sample containing buffer instead of lysate, but the difference was not statistically significant. Interestingly, the cell-free lysate from the CUP1his strain clearly suppressed the time-dependent increase in fluorescence. These results showed that the accumulated Cup1-His in the cell-free lysate from the CUP1his strain quenches NO, suggesting that Cup1 functions as an NO scavenger.
–
–
The expression level of CUP1 does not change under nitrosative stress conditions
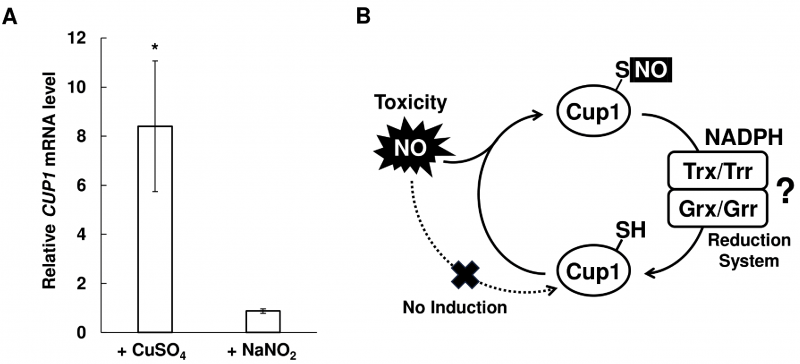
Genes involved in stress tolerance are often upregulated by the corresponding stress stimuli. Transcription of many genes encoding the proteins involved in NO detoxification, such as fHb and GSNOR, are induced by NO, although NO inhibits the copper- and Ace1-dependent induction of the CUP1 gene expression [36]. Therefore, we analyzed the transcriptional change of CUP1 in response to nitrosative stress (Figure 3A). We found that the CUP1 mRNA level was increased 8-fold in response to CuSO4 treatment, consistent with a previous study [37]. On the other hand, nitrosative stress induced by acidified nitrite treatment did not change the transcription level of CUP1, indicating that CUP1 expression does not respond to nitrosative stress. These results suggest that the Cup1-dependent NO tolerance mechanism functions as a constitutive NO tolerance system.
–
DISCUSSION
A previous report has shown that the overproduction of MT enhanced nitrosative stress tolerance and the MT-encoding gene was upregulated by RNS in mammalian cells [27]. However, it has not been clarified the functions of MT in yeast in relation to nitrosative stress. It has also been unclear whether MT expressed at the basal level or regulated by the endogenous expression system, not only in yeast but also in other organisms including mammalians. In this study, we showed that the yeast MT Cup1 under the physiological expression mechanism contributes to the nitrosative stress tolerance in S. cerevisiae cells, by scavenging NO. This is the first report not only to demonstrate the functions of the yeast MT related to NO, but also to indicate that MT with physiological expression regulation contributes to nitrosative stress tolerance.
–
Cysteine residues in proteins are susceptible to NO-dependent posttranslational modifications, such as S-nitrosylation and S-glutathionylation. Thus, MT is likely to be S-nitrosylated under nitrosative stress conditions, like other proteins with CXXC motifs. A previous study showed that MT was S-glutathionylated after nitrosative or oxidative stress treatment in the presence of GSH in mammalian cells [38]. It has also been reported that MT forms intramolecular disulfide bonds under oxidative stress conditions [24], suggesting that Cup1 forms disulfide bonds in it under nitrosative stress conditions, since NO functions as an oxidant. These results suggest that Cup1 is converted into the form with S-nitrosylation, S-glutathionylation, or an intramolecular disulfide bond in yeast cells in response to nitrosative stress. Whereas, A. nidulans nitrosothionein comprises MT-like motif and Trx/Trx reduction system reduces its S-nitrosylated cysteine residues to unmodified form. Therefore, some reduction mechanisms, including the Trx/Trr reduction system and/or glutaredoxin/glutaredoxin (Grx/Grr) reduction system, could function coordinately with Cup1 to regenerate the unmodified form of Cup1 by reducing these oxidative modifications (Figure 3B).
–
Our transcriptional analysis demonstrated that the expression level of CUP1 was not affected by nitrosative stress, which is similar to the finding that GCH2 is not induced by NO [14], and therefore Cup1 would function in a constitutive NO tolerance mechanism. In contrast, the expression of MT is induced by NO stimuli in mammalian cells [27], suggesting that a mammalian MT functions in an inducible defense system against nitrosative stress. The different regulations of MT among host species could be derived from the different amount of MT in the absence of stress stimuli in each organism at least partly. It is possible that the intracellular concentration of Cup1 is sufficient to fully scavenge NO without NO-dependent upregulation in yeast. The difference in the regulation mechanisms of MT can also account for the appropriate response time in yeasts and mammals. The constitutively functioning NO tolerance mechanism involving Cup1 could be necessary and efficient against acute nitrosative stress, which might occur in the living environment of yeast, since this mechanism does not require the time for transcription and translation. On the other hand, the inducible nitrosative stress tolerance mechanism by MT in mammalian cells would be an important factor to attenuate the toxicity of NO generated more slowly, because this inducible system requires the time for transcription and translation for the responsible proteins. Furthermore, the different response of MT to NO stimuli could be related with the particular mechanism by which nitrosative stress exerts its toxicity in the organism. In yeast, the reduced form of Cup1 is decreased by the oxidative modification induced by NO, since the protein level of Cup1 does not increase. Thus, transition metal ions originally bound to Cup1 could be released. However, the amount of transition metal released from MT under nitrosative stress conditions in mammalian cells should be limited more than yeast cells, because the MT protein level is increased in response to NO and thus the protein concentration of unmodified MT is still adequate to trap transition metal ions. Therefore, the disrupted homeostasis of transition metal would be one of the mechanisms involved in NO toxicity. Yeast cells might be more resistant to an increased concentration or disrupted homeostasis of transition metal ion than mammalian cells.
MATERIALS AND METHODS
Strains, plasmids, and media
S. cerevisiae BY4741 (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0) [39] was used as WT strain or a host strain to construct yeast mutant strains. BY4741 strain was transformed with a DNA fragment amplified by PCR using the plasmid pFA6a-natNT2 (EUROSCARF) as a template and the primers listed in Table 1 to construct the cup1Δ strain. The DNA fragment encoding Cup1 or Cup1-His was amplified by PCR using a chromosomal DNA of S. cerevisiae BY4741 strain as a template with the primers listed in Table 1 and inserted into the plasmid pAG425GPD-ccdB (Addgene) or pAG426GPD-ccdB (Addgene) by the BP and LR reactions of the Gateway technology (Invitrogen) following the manufacturer's protocol, generating pAG425GPD-CUP1, pAG426GPD-CUP1, pAG425GPD-CUP1-6HIS, or pAG426GPD-CUP1-6HIS. The resulting plasmids pAG425GPD-CUP1 and pAG426GPD-CUP1 were used to overexpress CUP1 in WT strain. The plasmids pAG425GPD-CUP1-6HIS and pAG426GPD-CUP1-6HIS were introduced into the cup1Δ strain to construct the CUP1his strain which overproduces Cup1-His. The plasmid pAG425GPD-ccdB and pAG426GPD-ccdB were introduced to WT or the cup1Δ strain as empty vectors. In order to compensate the remaining auxotrophic marker, pRS313-MET15 harboring HIS3 gene from Candida glabrata and MET15 gene from S. cerevisiae was introduced each strain [39].
–
Yeast cells were cultured at 25°C in SD medium containing 2% (wt/vol) glucose, 0.5% (wt/vol) ammonium sulfate, 0.17% (wt/vol) DifcoTM yeast nitrogen base without amino acids and ammonium sulfate (Becton, Dickinson), with 2% (wt/vol) agar if necessary. pH of SD medium was adjusted to 4.6 or 6.0.
–
Quantitative RT-PCR Analysis
Yeast cells cultured until the exponential growth phase in SD medium (pH 4.6) at 25°C were incubated in the absence or presence of 20 μM CuSO4 or 2 mM NaNO2 for 2 h. Harvested cells were disrupted using Multi-Beads Shocker (Yasui Kikai) with glass beads, and the total RNA was extracted with the RNeasy Mini Kit (QIAGEN) according to the manufacturer's instructions. cDNA was synthesized from the total RNA with the PrimeScript RT reagent Kit (Takara Bio). An RT-qPCR was performed using QuantStudio (Applied Biosystems) and SsoAdvanced Universal SYBR Green Supermix (Bio-Rad Laboratories) with the primer listed in Table 1. The ACT1 gene was used as a reference gene and the relative mRNA level was calculated by the ΔΔCt method. The relative expression level of CUP1 was calculated with the CUP1 mRNA level in WT strain or that in the untreated sample as 1.0 in order to compare the CUP1 expression level among strains or analyze the gene expression in response to stress stimuli, respectively.
–
Measurement of the intracellular NO level
Yeast cells grown until the exponential growth phase in SD medium (pH 6.0) at 25°C were treated with 15 μM DAF-FM DA at 25°C for 30 min and then exposed to 1 mM NOC-5 for 1 and 2 h, followed by FCM using BD Accuri C6 Flow Cytometer (Becton Dickinson). To evaluate the intracellular NO level, the relative fluorescence intensity was expressed as a percentage, which were calculated as follows: [(mean of fluorescence intensity at the indicated time point)/(mean of fluorescence intensity at 0 min after NOC-5 treatment)] × 100.
–
NO quenching assay
Yeast cells cultured until the log phase in SD medium (pH 6.0) at 25°C were suspended in Lysis buffer containing 20 mM Tris-HCl buffer (pH 7.4), 150 mM NaCl, and 5% (wt/vol) glycerol, followed by cell disruption using Multi-Beads Shocker (Yasui Kikai). Supernatant after centrifugation was used as a cell-free extract. For the crude purification with size exclusion mode, the cell-free extract was subjected to the ultrafiltration using Amicon® Ultra 0.5-ml Centrifugal Filters with 30 K and the flow through fractions were collected. Furthermore, in order to remove small compound in lysate and concentrate protein, the collected fractions were ultrafiltrated with Amicon® Ultra 0.5-ml Centrifugal Filters with 3 K in order to exchange the solvent to Lysis buffer, and the residual fractions were pooled and used as a cell-free lysate in the further analyses. Subsequently, the reaction mixture containing 20 mM Tris-HCl buffer (pH 7.4), 7 μM DAF-FM, 200 μM NOC-5, and the resultant cell-free lysate was incubated at room temperature and the fluorescence was monitored over time by a plate reader TriStar2 LB942 (Berthold Technologies) using an F485 excitation filter and an F535 emission filter. The linear slopes from 100 sec to 600 sec after the beginning of reaction was used to calculate the rate of fluorescence increase.
–
SDS-PAGE and Western blot analysis
Yeast cells grown until the exponential growth phase in SD medium (pH6.0) at 25°C were suspended in Lysis buffer with 2 mM phenylmethylsulfonyl fluoride and then disrupted by Multi-Beads Shocker (Yasui Kikai). Supernatant after centrifugation with the unified protein concentration was mixed with SDS sample buffer containing 100 mM Tris-HCl (pH 6.8), 20% glycerol, 4% SDS, 0.1% bromophenol blue, and 10% 2-mercaptethanol, and then boiled for 5 min to denature protein. The denatured samples were subjected to SDS-PAGE followed by CBB staining or immunoblotting using Anti-Histidine-Tagged Protein Mouse mAb (13/45/31/2) (Sigma-Aldrich) (anti-His antibody) and Anti-Mouse IgG (H + L), HRP Conjugate (Promega) (anti-mouse antibody). The visualization of immunoblot was performed using PierceTM ECL Plus Western Blotting Substrate (Thermo Fisher Scientific) and Image Quant LAS-4000 (Fujifilm).
REFERENCES
- Pacher P, Beckman JS, and Liaudet L (2007). Nitric oxide and peroxynitrite in health and disease. Physiol Rev 87(1): 315–424. 10.1152/physrev.00029.2006
- Wink DA, Hines HB, Cheng RYS, Switzer CH, Flores-Santana W, Vitek MP, Ridnour LA, and Colton CA (2011). Nitric oxide and redox mechanisms in the immune response. J Leukoc Biol 89(6): 873–891. 10.1189/jlb.1010550
- Allain A v., Hoang VT, Lasker GF, Pankey EA, Murthy SN, and Kadowitz PJ (2011). Role of nitric oxide in developmental biology in plants, bacteria, and man. Curr Top Pharmacol 15(2): 25–33. 24563585. 24563585
- Perez-Zoghbi JF, Bai Y, and Sanderson MJ (2010). Nitric oxide induces airway smooth muscle cell relaxation by decreasing the frequency of agonist-induced Ca2+ oscillations. J Gen Physiol 135(3): 247–259. 10.1085/jgp.200910365
- Nishimura A, Kawahara N, and Takagi H (2013). The flavoprotein Tah18-dependent NO synthesis confers high-temperature stress tolerance on yeast cells. Biochem Biophys Res Commun 430(1): 137–143. 10.1016/j.bbrc.2012.11.023
- Nasuno R, Aitoku M, Manago Y, Nishimura A, Sasano Y, and Takagi H (2014). Nitric oxide-mediated antioxidative mechanism in yeast through the activation of the transcription factor Mac1. PLoS One 9(11). 10.1371/journal.pone.0113788
- Nguyen T, Brunson D, Crespit CL, Penmant BW, Wishnok JS, and Tannenbaum SR (1992). DNA damage and mutation in human cells exposed to nitric oxide in vitro. Proc Natl Acad Sci USA 789(7): 3030-3034. 10.1073/pnas.89.7.3030
- Malawista SE, Montgomery RR, and van Blaricom G (1992). Evidence for reactive nitrogen intermediates in killing of staphylococci by human neutrophil cytoplasts. A new microbicidal pathway for polymorphonuclear leukocytes. J Clin Invest 90(2): 631–636. 10.1172/JCI115903
- Kaplan SS, Lancaster JR, Basford RE, and Simmons RL (1996). Effect of nitric oxide on staphylococcal killing and interactive effect with superoxide. Infect Immun 64(1): 69-76. 10.1128/iai.64.1.69-76.1996
- Kim SO, Orii Y, Lloyd D, Hughes MN, and Poole RK (1999). Anoxic function for the Escherichia coli flavohaemoglobin (Hmp): Reversible binding of nitric oxide and reduction to nitrous oxide. FEBS Lett 445(2–3): 389–394. 10.1016/S0014-5793(99)00157-X
- Liu L, Zeng M, Hausladen A, Heitman J, and Stamler JS (2000). Protection from nitrosative stress by yeast flavohemoglobin. Proc Natl Acad Sci USA 97(9): 4672–4676. 10.1073/pnas.090083597
- Wink DA, and Mitchell JB (1998). Chemical biology of nitric oxide: Insights into regulatory, cytotoxic, and cytoprotective mechanisms of nitric oxide. Free Radic Biol Med 25(4–5): 434–456. 10.1016/S0891-5849(98)00092-6
- Yoshikawa Y, Nasuno R, and Takagi H (2021). An NADPH-independent mechanism enhances oxidative and nitrosative stress tolerance in yeast cells lacking glucose-6-phosphate dehydrogenase activity. Yeast 38(7): 414–423. 10.1002/yea.3558
- Anam K, Nasuno R, and Takagi H (2020). A novel mechanism for nitrosative stress tolerance dependent on GTP cyclohydrolase II activity involved in riboflavin synthesis of yeast. Sci Rep 10(1): 1–10. 10.1038/s41598-020-62890-3
- Nikitovic D and Holmgren A (1996). S-nitrosoglutathione is cleaved by the thioredoxin system with liberation of glutathione and redox regulating nitric oxide. J Biol Chem 271(32): 19180–19185. 10.1074/jbc.271.32.19180
- Zhou S, Narukami T, Masuo S, Shimizu M, Fujita T, Doi Y, Kamimura Y, and Takaya N (2013). NO-inducible nitrosothionein mediates NO removal in tandem with thioredoxin. Nat Chem Biol 9(10): 657–663. 10.1038/nchembio.1316
- Olafson RW, McCubbin WD, and Kay CM (1988). Primary- and secondary-structural analysis of a unique prokaryotic metallothionein from a Synechococcus sp. cyanobacterium. Biochem J 251(3). 10.1042/bj2510691
- de Miranda JR, Thomas MA, Thurman DA, and Tomsett AB (1990). Metallothionein genes from the flowering plant Mimulus guttatus. FEBS Lett 260(2): 277–280. 10.1016/0014-5793(90)80122-Y
- Yutaka K, Christine B, Bertl V, and Jeremiash.R K (1976). Amino-acid sequence of equine renal metallothionein-IB. Proc Natl Acad Sci USA 73(10): 3413–3417. 10.1073/pnas.73.10.3413
- Kagi JH and Valee BL (1960). Metallothionein: a cadmium- and zinc-containing protein from equine renal cortex. J Biol Chem 235: 3460–3465. 10.1016/s0021-9258(18)64490-4
- Shaw CF, Savas MM, and Petering DH (1991). Ligand substitution and sulfhydryl reactivity of metallothionein. Methods Enzymol 205(C): 401–414. 10.1016/0076-6879(91)05122-C
- Thornalley PJ and Vašák M (1985). Possible role for metallothionein in protection against radiation-induced oxidative stress. Kinetics and mechanism of its reaction with superoxide and hydroxyl radicals. Biochim Biophys Acta Prot Struct Mol 827(1): 36–44. 10.1016/0167-4838(85)90098-6
- Misra RR, Hochadel JF, Smith GT, Cook JC, Waalkes MP, and Wink DA (1996). Evidence that nitric oxide enhances cadmium toxicity by displacing the metal from metallothionein. Chem Res Toxicol 9(1): 326–332. 10.1021/tx950109y
- Ruttkay-Nedecky B, Nejdl L, Gumulec J, Zitka O, Masarik M, Eckschlager T, Stiborova M, Adam V, and Kizek R (2013). The role of metallothionein in oxidative stress. Int J Mol Sci 14(3):6044–6066. 10.3390/ijms14036044
- Schwarz MA, Lazo JS, Yalowich JC, Allen WP, Whitmore M, Bergonia HA, Tzeng E, Billiar TR, Robbins PD, Lancaster JR, and Pitt BR (1995). Metallothionein protects against the cytotoxic and DNA-damaging effects of nitric oxide. Proc Natl Acad Sci USA 92(10): 4452–4456. 10.1073/pnas.92.10.4452
- Cai L, Klein JB, and Kang YJ (2000). Metallothionein inhibits peroxynitrite-induced DNA and lipoprotein damage. J Biol Chem 275(50): 38957–38960. 10.1074/jbc.C000593200
- Datta PK and Lianos EA (2006). Nitric oxide induces metallothionein-I gene expression in mesangial cells. Transl Res 148(4): 180–187. 10.1016/j.trsl.2006.04.002
- Fogel S and Welch JW (1982). Tandem gene amplification mediates copper resistance in yeast. Proc Natl Acad Sci USA 79(17): 5342–5346. 10.1073/pnas.79.17.5342
- Jon MH, David RE, and Dennis JT (1989). Copper-induced binding of cellular factors to yeast metallothionein upstream activation sequences. Proc Natl Acad Sci USA 86(1): 65–69. 10.1073/pnas.86.1.65
- Ecker DJ, Butt TR, Sternberg EJ, Neeper MP, Debouck C, Gorman JA, and Crooke ST (1986). Yeast metallothionein function in metal ion detoxification. J Biol Chem 261(36): 16895–16900. 10.1016/s0021-9258(19)75973-0
- Wegner S v., Sun F, Hernandez N, and He C (2011). The tightly regulated copper window in yeast. Chem Commun 47(9): 2571–2573. 10.1039/c0cc04292g
- Liu XD, and Thiele DJ (1996). Oxidative stress induces heat shock factor phosphorylation and HSF-dependent activation of yeast metallothionein gene transcription. Genes Dev 10(5): 592–603. 10.1101/gad.10.5.592
- Culotta VC, Howard WR, and Liu XF (1994). CRS5 encodes a metallothionein-like protein in Saccharomyces cerevisiae. J Biol Chem 269(41): 25295-25302. 10.1016/S0021-9258(18)47246-8
- Astuti RI, Watanabe D, and Takagi H (2016). Nitric oxide signaling and its role in oxidative stress response in Schizosaccharomyces pombe. Nitric Oxide 52: 29-40. 10.1016/j.niox.2015.11.001
- Lundberg JON, Weitzberg E, Lundberg JM, and Alving K (1994). Intragastric nitric oxide production in humans: Measurements in expelled air. Gut 35(11): 1543–1546. 10.1136/gut.35.11.1543
- Shinyashiki M, Chiang KT, Switzer CH, Gralla EB, Valentine JS, Thiele DJ, and Fukuto JM (2000). The interaction of nitric oxide (NO) with the yeast transcription factor Ace1: A model system for NO-protein thiol interactions with implications to metal metabolism. Proc Natl Acad Sci USA 97(6): 2491–2496. 10.1073/pnas.050586597
- Peña MMO, Koch KA, and Thiele DJ (1998). Dynamic regulation of copper uptake and detoxification genes in Saccharomyces cerevisiae. Mol Cell Biol 18(5): 2514–2523. 10.1128/mcb.18.5.2514
- Casadei M, Persichini T, Polticelli F, Musci G, and Colasanti M (2008). S-Glutathionylation of metallothioneins by nitrosative/oxidative stress. Exp Gerontol 43(5): 415–422. 10.1016/j.exger.2007.11.004
- Brachmann CB, Davies A, Cost GJ, Caputo E, Li J, Hieter P, and Boeke JD (1998). Designer deletion strains derived from Saccharomyces cerevisiae S288C: A useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14(2): 115–132. 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2
–
AUTHOR CONTRIBUTIONS
Y.Y., R.N. and H.T. conceived the study and designed the experiments. Y.Y. and R.N. performed the experiments. Y.Y., R.N., N.T., and H.T. analyzed the data and wrote the manuscript. All authors read and approved the final manuscript.
ACKNOWLEDGMENTS
This work was supported by grants for a Grant-in-Aid for Young Scientists (19K15808) from the Japan Society for the Promotion of Science (JSPS) to Y.Y., a Grant-in-Aid for Young Scientists (19K16129) from JSPS to R.N., a Grant-in-Aid for Scientific Research (C) (22K06084) from JSPS to R.N., and a Grant-in-Aid for Scientific Research (S) (19H05639), from JSPS to H.T.
COPYRIGHT
© 2023

Metallothionein Cup1 attenuates nitrosative stress in the yeast Saccharomyces cerevisiae by Yoshikawa et al. is licensed under a Creative Commons Attribution 4.0 International License.