Microreviews:
Microbial Cell, Vol. 3, No. 2, pp. 89 - 91; doi: 10.15698/mic2016.02.479
Location, location, location. Salmonella senses ethanolamine to gauge distinct host environments and coordinate gene expression
Department of Microbiology, Immunology, and Cancer Biology, University of Virginia School of Medicine, Charlottesville, Virginia, U.S.A.
Keywords: Salmonella, ethanolamine, macrophages, GI tract, metabolism.
Received originally: 23/12/2015 Accepted: 06/01/2016
Published: 18/01/2016
Correspondence:
Melissa M. Kendall, melissakendall@virginia.edu
Conflict of interest statement: The authors declare that no competing interest exists.
Please cite this article as: Christopher J. Anderson and Melissa M. Kendall (2016). Location, location, location. Salmonella senses ethanolamine to gauge distinct host environments and coordinate gene expression. Microbial Cell 3(2): 89-91.
Chemical and nutrient signaling mediate all cellular processes, ensuring survival in response to changing environmental conditions. Ethanolamine is a component of phosphatidylethanolamine, a major phospholipid of mammalian and bacterial cell membranes. Ethanolamine is abundant in the gastrointestinal (GI) tract from dietary sources as well as from the normal turnover of intestinal epithelial and bacterial cells in the gut. Additionally, mammalian cells maintain intracellular ethanolamine concentrations through low and high-affinity uptake systems and the internal recycling of phosphatidylethanolamine; therefore, ethanolamine is ubiquitous throughout the mammalian host. Although ethanolamine has profound signaling activity within mammalian cells by modulating inflammatory responses and intestinal physiology, ethanolamine is best appreciated as a nutrient for bacteria that supports growth. In our recent work (Anderson, et al. PLoS Pathog (2015), 11: e1005278), we demonstrated that Salmonella enterica serovar Typhimurium (Salmonella) exploits ethanolamine signaling to adapt to distinct host environments to precisely coordinate expression of genes encoding metabolism and virulence, which ultimately enhances disease progression.
–
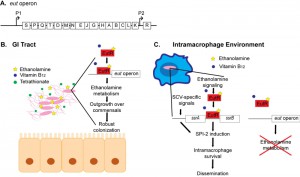
Gram-negative and Gram-positive bacteria encode the ethanolamine utilization (eut) operon. This operon has been most widely studied in the Enterobacteriaceae, and several research groups established that this operon contains 17 genes encoding enzymes and a microcompartment necessary for the breakdown of ethanolamine (Figure 1A). The eut operon also encodes the transcriptional regulator EutR. Genetic studies by Roof and Roth showed that EutR is constitutively expressed at low levels from the P2 promoter, and in the presence of ethanolamine and the cofactor vitamin B12, EutR induces expression of the entire eut operon. Previously, we used biochemical approaches and demonstrated that EutR directly senses ethanolamine and binds to the eutS (P1) promoter to activate transcription (Figure 1A). Additionally, we reported that enterohemorrhagic Escherichia coli exploits ethanolamine signaling through EutR to activate expression of virulence genes in vitro, which established the importance of ethanolamine as a bacterial signaling molecule. However, the question remained whether ethanolamine signaling influences bacterial virulence in vivo.
To address this, we investigated ethanolamine signaling in Salmonella pathogenesis. Salmonella is a food-borne pathogen that causes acute gastroenteritis. Although Salmonella infections are typically limited to the GI tract, a subset of infected individuals will develop systemic infections that can be fatal. Salmonella infection presents as intestinal outgrowth, penetration of the epithelial barrier, and subsequent uptake by macrophages, where Salmonella is ultimately trafficked to secondary lymphoid organs such as the spleen. To address whether ethanolamine signaling impacted Salmonella disease progression during infection, we performed competition experiments and examined recovery of wild type (WT) Salmonella, the ΔeutR strain that cannot respond to ethanolamine or the ΔeutB strain that cannot metabolize ethanolamine, from the intestinal contents, the colon, and the spleen. After orogastric infection of streptomycin-treated mice, the ∆eutR and ∆eutB strains were recovered in significantly lower numbers compared to WT from intestinal contents, indicating that ethanolamine metabolism is critical for Salmonella colonization of the GI tract. These results agreed with previous findings by the Bäumler lab that reported that Salmonella respires ethanolamine in conjunction with the inflammation-derived electron acceptor tetrathionate to outgrow the resident microbiota and establish infection. Significantly, examination of splenic tissue revealed that the ∆eutB strain was only slightly attenuated in dissemination to the spleen compared to WT, whereas the ∆eutR strain displayed a more robust disadvantage in dissemination to the spleen. To distinguish between ethanolamine metabolism-dependent and -independent roles for EutR in vivo, we directly competed the ∆eutR and the ∆eutB strains. No differences in the numbers of recovered bacteria were measured after 2 days post infection (dpi) in the intestinal contents, indicating that at this early stage and site of infection EutR functions primarily to direct ethanolamine metabolism. However, at 4 dpi the ∆eutR strain was significantly outcompeted by the ∆eutB strain in the intestinal contents. This time point is consistent with progression to systemic infection, and in agreement, the ∆eutR strain was recovered in significantly lower numbers compared to the ∆eutB strain from the spleen. These data indicated that the role of EutR during infection is dynamic and extends beyond promoting metabolism.
–
Intramacrophage survival and replication is a major means by which Salmonella disseminates, and we focused on the role of ethanolamine signaling on this aspect of Salmonella pathogenesis. To determine how EutR functions to promote dissemination, we assessed Salmonella survival within RAW and primary macrophages. Following macrophage infection, the ΔeutR strain was recovered at significantly lower numbers compared to WT or the ΔeutB strains, indicating that ethanolamine signaling through EutR enhances Salmonella survival within macrophages, in a manner independent from promoting metabolism.
–
Based on these findings, we hypothesized that EutR regulates expression of the Salmonella pathogenicity island (SPI)-2. SPI-2 contains four operons that encode a type three secretion system and effectors. Additionally, SPI-2 encodes a two component system (TCS), SsrAB, in which SsrA is the sensor kinase and SsrB is the response regulator that directly activates SPI-2 expression. Extensive research has established that SPI-2 expression is restricted to the intracellular environment and is essential for intramacrophage survival. Expression of ssrB and downstream SPI-2 operons was significantly decreased in the ΔeutR strain compared to WT Salmonella during macrophage infection, indicating that ethanolamine signaling plays an important role in Salmonella virulence regulation. Interestingly, although ssrB expression was decreased in the ΔeutR strain, EutR did not modulate ssrA expression. The ssrAB operon contains two promoters, one immediately upstream of ssrA and a second promoter immediately upstream of ssrB, which allow for ssrB expression independent of SsrA. In vitro assays using purified EutR in conjunction with in vivo assays revealed that EutR directly binds the ssrB promoter to activate SPI-2 expression. These findings indicate a more extensive role for EutR in Salmonella pathogenesis than previously appreciated, as a direct regulator of virulence traits.
–
Control of SPI-2 expression is complex and also includes additional regulatory systems, including the EnvZ-OmpR and PhoP-PhoQ TCSs. These TCSs respond to stimuli in the Salmonella – containing vacuole (SCV) (a modified phagosome within which Salmonella replicates in macrophages) to activate SPI-2 expression. Our findings suggest a model in which EutR-dependent activation of ssrB enables Salmonella to incorporate ethanolamine, an abundant and integral component of host cell membranes, with SCV-specific signals to efficiently and temporally regulate SPI-2 expression. Further investigation is required to understand the interplay between EutR and the EnvZ-OmpR/PhoP-PhoQ systems in promoting SPI-2 expression.
–
To further elucidate the dynamics of EutR-associated signaling, we assessed expression of the eut operon in WT Salmonella during macrophage infection. Expression of eutR was highly induced within macrophages compared to Salmonella grown in the absence of macrophages, confirming that the intramacrophage environment is sufficient to activate EutR-associated signaling. Notably, eutR expression was most highly induced at 3 hpi, and expression levels decreased as infection progressed. These data suggest that ethanolamine signaling is important for the initial adaptation of Salmonella to the intramacrophage environment. Surprisingly, despite robust eutR expression, we did not measure an induction of the eut operon (as determined by measuring eutS expression) within macrophages.
–
To confirm these findings within the complexities of the in vivo environment, we performed competition infections using an intraperitoneal infection model, which specifically examines SPI-2 mediated systemic disease. We assessed recovery of the ΔeutR and ΔeutB strains from the peritoneal cavity as well as within peritoneal phagocytes. The ΔeutR and ΔeutB strains were equally fit in the peritoneal cavity, which contained primarily extracellular Salmonella; however, the ΔeutR strain was significantly outcompeted by the ΔeutB strain within the resident phagocyte population. Subsequently, we performed single infections with WT and the ∆eutR strains and measured gene expression from Salmonella recovered from spleens. In agreement with the macrophage data, expression of ssrB was significantly decreased in the ∆eutR strain compared to WT. Moreover, although eutR expression was significantly induced in WT Salmonella, eutS expression was below detection limits. Altogether, these findings highlight that EutR directs gene expression specifically according to a particular environment.
–
Pathogens rely on precise and coordinated gene expression to successfully grow within a host and cause disease. Our recent work highlights a sophisticated mechanism in which Salmonella exploits ethanolamine to spatiotemporally control expression of genes encoding metabolism and virulence. During the initial stages of Salmonella infection, EutR activates expression of the eut operon. This promotes robust growth of Salmonella and establishment of infection (Figure 1B). In the intracellular environment where ethanolamine metabolism in not energetically favorable, we hypothesize that EutR preferentially targets genes required for macrophage adaptation (Figure 1C). How EutR differentially promotes gene expression according to location within a host is an ongoing area of research in our lab. Genes encoding ethanolamine metabolism are widely distributed among bacteria. Therefore, ethanolamine signaling may be a conserved strategy used by diverse pathogens to recognize and adapt to distinct host environments.
ACKNOWLEDGMENTS
This work was funded by the National Institutes of Health, National Institute of Allergy and Infectious Diseases (AI118732) to MMK and the training grant (5T32AI007046) to CJA. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
COPYRIGHT
© 2016

Location, location, location. Salmonella senses ethanolamine to gauge distinct host environments and coordinate gene expression by Christopher J. Anderson and Melissa M. Kendall is licensed under a Creative Commons Attribution 4.0 International License.