Editorial:
Microbial Cell, Vol. 1, No. 4, pp. 110 - 114; doi: 10.15698/mic2014.04.142
Metabolites in aging and autophagy
1 Institute of Molecular Biosciences, University of Graz, Humboldtstrasse 50, 8010 Graz, Austria.
2 Division of Endocrinology and Metabolism, Dept. of Internal Medicine, Medical University of Graz, Auenbruggerplatz 15, 8036 Graz, Austria.
# These authors contributed equally to this work.
Keywords: autophagy, aging, metabolism, acetyl-CoA, polyamines, amino acids.
Received originally: 21/03/2014 Received in revised form: 04/04/2014
Accepted: 04/04/2014
Published: 07/04/2014
Correspondence:
Frank Madeo, Humboldtstrasse 50; 8010 Graz, Austria frank.madeo@uni-graz.at
Conflict of interest statement: The authors declare no conflict of interest.
Please cite this article as: Sabrina Schroeder, Andreas Zimmermann, Didac Carmona-Gutierrez, Tobias Eisenberg, Christoph Ruckenstuhl, Aleksandra Andryushkova, Tobias Pendl, Alexandra Harger and Frank Madeo (2014). Metabolites in aging and autophagy. Microbial Cell 1(4): 110-114.
INTRODUCTION
Autophagy, the main lysosomal degradative machinery, plays a major role in maintaining cellular homeostasis and thus a healthy state in an organism. This process recycles unnecessary or damaged material, therefore, not only providing nutrients to maintain vital cellular functions in times of starvation but also eliminating potentially harmful cellular material [1]. Importantly, the autophagic rate declines with increasing age [2][3], suggesting a functional correlation between aging and autophagy. Indeed, the deregulation of autophagy is involved in the onset of various age-related diseases such as cancer, cardiomyopathy, type II diabetes, and neurodegeneration [4]. Until recently, aging was regarded as an unregulated and inescapable consequence of the accumulation of incidental damage in macromolecules and/or organelles. However, the discovery of multiple ways to extend the lifespan in a variety of different model organisms, e.g., by genetic and pharmacological means, developed the formulation of alternative aging theories that consider aging as a molecular program [5]. Indeed, the last years have provided important insights into the networks that control aging and have thus highlighted the interconnected nature of aging and various cellular processes. For instance, the process of aging is intimately coupled to metabolic processes [6], in particular to energy metabolism and nutrient availability. Nevertheless, specific metabolites that affect aging and autophagy remain poorly described.
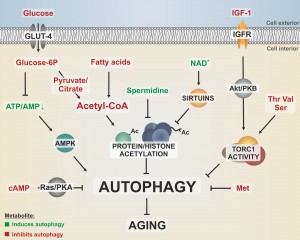
NUTRIENT AVAILABILITY CONTROLS AUTOPHAGY AND AGING VIA ENERGY METABOLITES
As sensors of the current environmental status, nutrient signaling pathways represent central aging regulators. For instance, individual interventions in the insulin/insulin-like growth factor 1 (IGF-1), Ras, protein kinase A (PKA), target of rapamycin (Tor), or protein kinase B (SCH9/Akt) pathways have been shown to extend lifespan in various organisms, including mammals [7][8][9]. Caloric restriction (CR) requires autophagy for lifespan extension [10][11][12] and CR-mediated autophagy induction follows molecular pathways that are shared with those known to affect aging, such as Tor, SCH9/Akt, or IGF-1 [13]. Furthermore, the AMP-activated protein kinase (AMPK) serves as a metabolic radar sensing changes in the AMP/ATP ratio and is conserved in the majority of eukaryotic species, and has also been established as a checkpoint for growth control and autophagy regulation [14]. Consistently, several studies have revealed a connection between the AMP/ATP ratio, autophagic flux rates, senescence, and disease [15][16]. Noteworthy, early studies on rat hepatocytes also suggested that the execution of autophagy depends on energy availability since inhibition of ATP production stalls autophagic flux [17]. Other pivotal energy sources like butyrate, an essential energy component in the colon, and second messengers such as cAMP, which might also be implicated in Ras/PKA-mediated lifespan modulation in various organisms, were identified as potential autophagy mediators [18][19]. This argues for a decisive function of nutrient signaling and energy metabolites during aging and its associated processes.
THE ENERGY METABOLITE ACETYL-CoA SUPPRESSES AUTOPHAGY VIA PROTEIN ACETYLATION
In the yeast S cerevisiae, glucose is the preferred carbon source for fueling energy metabolism and its fermentation produces acetate and ethanol that is used for subsequent respiration after the diauxic shift. Both intermediates are released into the medium and have been attributed a role in limiting yeast chronological life span [20], but considered a pro-aging factor specific for yeast [21]. However, acetate is an important metabolite that is involved in central processes such as acetyl-Coenzyme A (acetyl-CoA) production. Thus, its metabolic fate rather than a simply extracellular toxicity may (at least in part) be responsible for its impact on lifespan. Consequently, acetate metabolism may also contribute to aging in higher eukaryotes, possibly via its impact on protein acetylation through acetyl-CoA generation and subsequent control of cellular function. Energy metabolites that can derive in the production of acetyl-CoA, such as citrate, pyruvate and fatty acids, were shown to be deregulated in senescence-accelerated mice [22]. We could recently show that (nucleo-)cytosolic acetyl-CoA, in fact, serves as a modulator of longevity, suppressing starvation and age-associated autophagy in a variety of phyla [23][24]. This function might be due to the fact that acetyl-CoA is the only donor for acetylation reactions and both, protein acetylation and epigenetic chromatin modifications, have repeatedly been linked to the regulation of aging and autophagy [23][25][26]. For instance, the highly conserved protein family of NAD+-dependent histone deacetylases and ADP ribosylases (sirtuins) has been connected to aging modulation [27][28] and lifespan extension upon CR [29]. Activation of histone deacetylases, such as sirtuins, has therefore been extensively studied for its capability to combat aging or age-associated pathologies. Interestingly, the lifespan-extending effects of sirtuin activation by CR or pharmacological interventions depend on the induction of autophagy [11]. The dependency on NAD+ as a cofactor and the tight connections between sirtuins, longevity and autophagy induction have led to the hypothesis that sirtuins act as metabolic sensors that promote mitochondrial maintenance [30]. Notably, nicotinamide metabolism has become an intensively investigated target for drug discovery against a variety of human diseases, including age-associated pathologies such as cancer or neurodegeneration [31].
POLYAMINE METABOLISM LEAVES A REGULATORY FINGERPRINT AT HISTONE MODIFICATION SITES
The levels of polyamines, a class of ubiquitously occurring small basic polycations, decline with progressing age in various organisms, including humans [32], yeast [25] and plants [33]. Intriguingly, external application of a specific polyamine (spermidine) counteracts cell death during aging and improves the lifespan of yeast, flies, worms, and human immune cells in an autophagy-dependent manner [25]. Furthermore, it causes a reduction of oxidative stress in mice. In addition, more recent studies also suggest longevity-promoting effects in mammals [34]. Importantly, spermidine treatment appears to be associated with both histone hypoacetylation caused by inhibition of histone acetyl transferases [25] and deacetylation of cytosolic proteins [35]. However, it has also been hypothesized that polyamines influence histone acetylation in dependence of the histones’ own acetylation status [36] and massive polyamine catabolism has even been shown to deplete the acetylation co-factor acetyl-CoA [37]. Possibly, the involvement of spermidine in the biosynthesis of the methyl-group donor S-adenosyl-methionine (SAM) [38] could also culminate in regulatory methylation reactions, such as chromatin silencing. Thus, polyamines may influence chromatin structure and protein acetylation via multiple mechanisms. Given the aforementioned contribution of acetyl-CoA to aging and autophagy modulation, it is tempting to speculate that spermidine functions include a down-titration of the intracellular acetyl-CoA pool and thus a rearrangement of the metabolic state.
SPECIFIC AMINO ACID STARVATION INDUCES AUTOPHAGY AND PROLONGS LIFESPAN
In line with the vast importance of nutrient signaling during aging, amino acid metabolism has an important impact on eukaryotic aging and its related diseases. The levels of specific amino acids like tryptophane, methionine, arginine, or leucine has often been suggested to (positively or negatively) influence the autophagy pathway and impact aging in different eukaryotes [39][40][41][42]. Indeed, we could recently demonstrate that limitation of the amino acid methionine enhances yeast chronological lifespan. Intriguingly, this lifespan extension requires autophagy-dependent vacuolar acidification [43]. In the same line, vacuolar acidification also elongates replicative lifespan of yeast, where it protects mitochondria most probably via an improvement of the vacuolar amino acid storage function [44]. In yeast, serine, threonine and valine promote cellular senescence probably via activation of the Sch9/TOR pathway and subsequent inhibition of the protein kinase Rim15p, which guides anti-aging stress response pathways [45]. Altogether, these examples suggest that the intake or limitation of specific amino acids is a determining factor during aging, though the mechanistic specificities are expected to be complexly regulated, especially at the organismal level.
CONCLUSION
A variety of potential metabolic controllers of autophagy and health span have already been proposed. However, precise strategies to target the correlating pathways (e.g., by nutrition patterns) remain to be elucidated in more detail. For example, it would be of great interest to determine if special diets that include the limitation of (defined) amino acids or the uptake of certain polyamines, like spermidine, influence the metabolism towards improved cellular conditions during aging. It also remains elusive how certain diets may affect the microbiome and in turn impact the levels of certain metabolites that have been shown to regulate cellular fitness, such as citrate, pyruvate, butyrate, or acetate. The investigation of metabolites as powerful rheostats in aging and autophagy is supported by the improvement of technologies that have opened up new possibilities to detect and trace even small molecules in vitro and in vivo. This might bring up metabolomics as a future trend for aging analyses [46]. Hopefully, the findings on the impact of metabolism on aging will culminate in amended dietary guidelines that would make eating the tastiest of all medicines.
References
- G. Kroemer, G. Mariño, and B. Levine, "Autophagy and the Integrated Stress Response", Molecular Cell, vol. 40, pp. 280-293, 2010. http://dx.doi.org/10.1016/j.molcel.2010.09.023
- A.M. Cuervo, "Autophagy and aging: keeping that old broom working", Trends in Genetics, vol. 24, pp. 604-612, 2008. http://dx.doi.org/10.1016/j.tig.2008.10.002
- A. Simonsen, R.C. Cumming, A. Brech, P. Isakson, D.R. Schubert, and K.D. Finley, "Promoting basal levels of autophagy in the nervous system enhances longevity and oxidant resistance in adult Drosophila.", Autophagy, 2007. http://www.ncbi.nlm.nih.gov/pubmed/18059160
- B. Levine, and G. Kroemer, "Autophagy in the Pathogenesis of Disease", Cell, vol. 132, pp. 27-42, 2008. http://dx.doi.org/10.1016/j.cell.2007.12.018
- T.C. Goldsmith, "On the programmed/non-programmed aging controversy", Biochemistry (Moscow), vol. 77, pp. 729-732, 2012. http://dx.doi.org/10.1134/S000629791207005X
- R.H. Houtkooper, R.W. Williams, and J. Auwerx, "Metabolic Networks of Longevity", Cell, vol. 142, pp. 9-14, 2010. http://dx.doi.org/10.1016/j.cell.2010.06.029
- V.D. Longo, "Mutations in signal transduction proteins increase stress resistance and longevity in yeast, nematodes, fruit flies, and mammalian neuronal cells", Neurobiology of Aging, vol. 20, pp. 479-486, 1999. http://dx.doi.org/10.1016/S0197-4580(99)00089-5
- M. Tatar, A. Kopelman, D. Epstein, M. Tu, C. Yin, and R.S. Garofalo, "A Mutant Drosophila Insulin Receptor Homolog That Extends Life-Span and Impairs Neuroendocrine Function", Science, vol. 292, pp. 107-110, 2001. http://dx.doi.org/10.1126/science.1057987
- M. Kaeberlein, R.W. Powers, K.K. Steffen, E.A. Westman, D. Hu, N. Dang, E.O. Kerr, K.T. Kirkland, S. Fields, and B.K. Kennedy, "Regulation of Yeast Replicative Life Span by TOR and Sch9 in Response to Nutrients", Science, vol. 310, pp. 1193-1196, 2005. http://dx.doi.org/10.1126/science.1115535
- K. Jia, and B. Levine, "Autophagy is required for dietary restriction-mediated life span extension in C. elegans.", Autophagy, 2007. http://www.ncbi.nlm.nih.gov/pubmed/17912023
- E. Morselli, M.C. Maiuri, M. Markaki, E. Megalou, A. Pasparaki, K. Palikaras, A. Criollo, L. Galluzzi, S.A. Malik, I. Vitale, M. Michaud, F. Madeo, N. Tavernarakis, and G. Kroemer, "Caloric restriction and resveratrol promote longevity through the Sirtuin-1-dependent induction of autophagy", Cell Death & Disease, vol. 1, pp. e10-e10, 2010. http://dx.doi.org/10.1038/cddis.2009.8
- E.A. Minina, V. Sanchez‐Vera, P.N. Moschou, M.F. Suarez, E. Sundberg, M. Weih, and P.V. Bozhkov, "Autophagy mediates caloric restriction‐induced lifespan extension in Arabidopsis", Aging Cell, vol. 12, pp. 327-329, 2013. http://dx.doi.org/10.1111/acel.12048
- D. Rubinsztein, G. Mariño, and G. Kroemer, "Autophagy and Aging", Cell, vol. 146, pp. 682-695, 2011. http://dx.doi.org/10.1016/j.cell.2011.07.030
- M.M. Mihaylova, and R.J. Shaw, "The AMPK signalling pathway coordinates cell growth, autophagy and metabolism", Nature Cell Biology, vol. 13, pp. 1016-1023, 2011. http://dx.doi.org/10.1038/ncb2329
- Z. Yang, H. Fujii, S.V. Mohan, J.J. Goronzy, and C.M. Weyand, "Phosphofructokinase deficiency impairs ATP generation, autophagy, and redox balance in rheumatoid arthritis T cells", Journal of Experimental Medicine, vol. 210, pp. 2119-2134, 2013. http://dx.doi.org/10.1084/jem.20130252
- D.A. Kubli, and �.B. Gustafsson, "Cardiomyocyte health: adapting to metabolic changes through autophagy", Trends in Endocrinology & Metabolism, vol. 25, pp. 156-164, 2014. http://dx.doi.org/10.1016/j.tem.2013.11.004
- P.J.A.M. PLOMP, E.J. WOLVETANG, A.K. GROEN, A.J. MEIJER, P.B. GORDON, and P.O. SEGLEN, "Energy dependence of autophagic protein degradation in isolated rat hepatocytes", European Journal of Biochemistry, vol. 164, pp. 197-203, 1987. http://dx.doi.org/10.1111/j.1432-1033.1987.tb11011.x
- I. Holen, P.B. Gordon, P.E. Strømhaug, and P.O. Seglen, "Role of cAMP in the Regulation of Hepatocytic Autophagy", European Journal of Biochemistry, vol. 236, pp. 163-170, 1996. http://dx.doi.org/10.1111/j.1432-1033.1996.00163.x
- D. Donohoe, N. Garge, X. Zhang, W. Sun, T. O'Connell, M. Bunger, and S. Bultman, "The Microbiome and Butyrate Regulate Energy Metabolism and Autophagy in the Mammalian Colon", Cell Metabolism, vol. 13, pp. 517-526, 2011. http://dx.doi.org/10.1016/j.cmet.2011.02.018
- M.G. Mirisola, and V.D. Longo, "Acetic acid and acidification accelerate chronological and replicative aging in yeast", Cell Cycle, vol. 11, pp. 3532-3533, 2012. http://dx.doi.org/10.4161/cc.22042
- W.C. Burhans, and M. Weinberger, "Acetic acid effects on aging in budding yeast: Are they relevant to aging in higher eukaryotes?", Cell Cycle, vol. 8, pp. 2300-2302, 2009. http://dx.doi.org/10.4161/cc.8.14.8852
- N. Jiang, X. Yan, W. Zhou, Q. Zhang, H. Chen, Y. Zhang, and X. Zhang, "NMR-Based Metabonomic Investigations into the Metabolic Profile of the Senescence-Accelerated Mouse", Journal of Proteome Research, vol. 7, pp. 3678-3686, 2008. http://dx.doi.org/10.1021/pr800439b
- T. Eisenberg, S. Schroeder, A. Andryushkova, T. Pendl, V. Küttner, A. Bhukel, G. Mariño, F. Pietrocola, A. Harger, A. Zimmermann, T. Moustafa, A. Sprenger, E. Jany, S. Büttner, D. Carmona-Gutierrez, C. Ruckenstuhl, J. Ring, W. Reichelt, K. Schimmel, T. Leeb, C. Moser, S. Schatz, L. Kamolz, C. Magnes, F. Sinner, S. Sedej, K. Fröhlich, G. Juhasz, T. Pieber, J. Dengjel, S.J. Sigrist, G. Kroemer, and F. Madeo, "Nucleocytosolic Depletion of the Energy Metabolite Acetyl-Coenzyme A Stimulates Autophagy and Prolongs Lifespan", Cell Metabolism, vol. 19, pp. 431-444, 2014. http://dx.doi.org/10.1016/j.cmet.2014.02.010
- G. Mariño, F. Pietrocola, T. Eisenberg, Y. Kong, S. Malik, A. Andryushkova, S. Schroeder, T. Pendl, A. Harger, M. Niso-Santano, N. Zamzami, M. Scoazec, S. Durand, D. Enot, �. Fernández, I. Martins, O. Kepp, L. Senovilla, C. Bauvy, E. Morselli, E. Vacchelli, M. Bennetzen, C. Magnes, F. Sinner, T. Pieber, C. López-Otín, M. Maiuri, P. Codogno, J. Andersen, J. Hill, F. Madeo, and G. Kroemer, "Regulation of Autophagy by Cytosolic Acetyl-Coenzyme A", Molecular Cell, vol. 53, pp. 710-725, 2014. http://dx.doi.org/10.1016/j.molcel.2014.01.016
- T. Eisenberg, H. Knauer, A. Schauer, S. Büttner, C. Ruckenstuhl, D. Carmona-Gutierrez, J. Ring, S. Schroeder, C. Magnes, L. Antonacci, H. Fussi, L. Deszcz, R. Hartl, E. Schraml, A. Criollo, E. Megalou, D. Weiskopf, P. Laun, G. Heeren, M. Breitenbach, B. Grubeck-Loebenstein, E. Herker, B. Fahrenkrog, K. Fröhlich, F. Sinner, N. Tavernarakis, N. Minois, G. Kroemer, and F. Madeo, "Induction of autophagy by spermidine promotes longevity", Nature Cell Biology, vol. 11, pp. 1305-1314, 2009. http://dx.doi.org/10.1038/ncb1975
- S. Imai, C.M. Armstrong, M. Kaeberlein, and L. Guarente, "Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase", Nature, vol. 403, pp. 795-800, 2000. http://dx.doi.org/10.1038/35001622
- P. Fabrizio, C. Gattazzo, L. Battistella, M. Wei, C. Cheng, K. McGrew, and V.D. Longo, "Sir2 Blocks Extreme Life-Span Extension", Cell, vol. 123, pp. 655-667, 2005. http://dx.doi.org/10.1016/j.cell.2005.08.042
- M. Kaeberlein, M. McVey, and L. Guarente, "The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms.", Genes & development, 1999. http://www.ncbi.nlm.nih.gov/pubmed/10521401
- S. Lin, P. Defossez, and L. Guarente, "Requirement of NAD and SIR2 for Life-Span Extension by Calorie Restriction in Saccharomyces cerevisiae", Science, vol. 289, pp. 2126-2128, 2000. http://dx.doi.org/10.1126/science.289.5487.2126
- M.N. Sack, and T. Finkel, "Mitochondrial Metabolism, Sirtuins, and Aging", Cold Spring Harbor Perspectives in Biology, vol. 4, pp. a013102-a013102, 2012. http://dx.doi.org/10.1101/cshperspect.a013102
- J.A. Khan, F. Forouhar, X. Tao, and L. Tong, "Nicotinamide adenine dinucleotide metabolism as an attractive target for drug discovery", Expert Opinion on Therapeutic Targets, vol. 11, pp. 695-705, 2007. http://dx.doi.org/10.1517/14728222.11.5.695
- G. Scalabrino, and M.E. Ferioli, "Polyamines in mammalian ageing: an oncological problem, too? A review.", Mechanisms of ageing and development, 1984. http://www.ncbi.nlm.nih.gov/pubmed/6384679
- R. Kaur-Sawhney, L.M. Shih, H.E. Flores, and A.W. Galston, "Relation of polyamine synthesis and titer to aging and senescence in oat leaves.", Plant physiology, 1982. http://www.ncbi.nlm.nih.gov/pubmed/16662218
- K. Soda, Y. Dobashi, Y. Kano, S. Tsujinaka, and F. Konishi, "Polyamine-rich food decreases age-associated pathology and mortality in aged mice", Experimental Gerontology, vol. 44, pp. 727-732, 2009. http://dx.doi.org/10.1016/j.exger.2009.08.013
- E. Morselli, G. Mariño, M.V. Bennetzen, T. Eisenberg, E. Megalou, S. Schroeder, S. Cabrera, P. Bénit, P. Rustin, A. Criollo, O. Kepp, L. Galluzzi, S. Shen, S.A. Malik, M.C. Maiuri, Y. Horio, C. López-Otín, J.S. Andersen, N. Tavernarakis, F. Madeo, and G. Kroemer, "Spermidine and resveratrol induce autophagy by distinct pathways converging on the acetylproteome", Journal of Cell Biology, vol. 192, pp. 615-629, 2011. http://dx.doi.org/10.1083/jcb.201008167
- B. Liu, A. Sutton, and R. Sternglanz, "A Yeast Polyamine Acetyltransferase", Journal of Biological Chemistry, vol. 280, pp. 16659-16664, 2005. http://dx.doi.org/10.1074/jbc.M414008200
- K. Kee, B.A. Foster, S. Merali, D.L. Kramer, M.L. Hensen, P. Diegelman, N. Kisiel, S. Vujcic, R.V. Mazurchuk, and C.W. Porter, "Activated Polyamine Catabolism Depletes Acetyl-CoA Pools and Suppresses Prostate Tumor Growth in TRAMP Mice", Journal of Biological Chemistry, vol. 279, pp. 40076-40083, 2004. http://dx.doi.org/10.1074/jbc.M406002200
- M. Sauter, B. Moffatt, M. Saechao, R. Hell, and M. Wirtz, "Methionine salvage and S-adenosylmethionine: essential links between sulfur, ethylene and polyamine biosynthesis", Biochemical Journal, vol. 451, pp. 145-154, 2013. http://dx.doi.org/10.1042/BJ20121744
- E.L. Dillon, "Nutritionally essential amino acids and metabolic signaling in aging", Amino Acids, vol. 45, pp. 431-441, 2012. http://dx.doi.org/10.1007/s00726-012-1438-0
- N. Orentreich, J.R. Matias, A. DeFelice, and J.A. Zimmerman, "Low methionine ingestion by rats extends life span.", The Journal of nutrition, 1993. http://www.ncbi.nlm.nih.gov/pubmed/8429371
- A.T. van der Goot, and E.A. Nollen, "Tryptophan metabolism: entering the field of aging and age-related pathologies", Trends in Molecular Medicine, vol. 19, pp. 336-344, 2013. http://dx.doi.org/10.1016/j.molmed.2013.02.007
- J.P. Aris, A.L. Alvers, R.A. Ferraiuolo, L.K. Fishwick, A. Hanvivatpong, D. Hu, C. Kirlew, M.T. Leonard, K.J. Losin, M. Marraffini, A.Y. Seo, V. Swanberg, J.L. Westcott, M.S. Wood, C. Leeuwenburgh, and W.A. Dunn, "Autophagy and leucine promote chronological longevity and respiration proficiency during calorie restriction in yeast", Experimental Gerontology, vol. 48, pp. 1107-1119, 2013. http://dx.doi.org/10.1016/j.exger.2013.01.006
- C. Ruckenstuhl, C. Netzberger, I. Entfellner, D. Carmona-Gutiérrez, T. Kickenweiz, S. Stekovic, C. Gleixner, C. Smidt, L. Klug, A. Sorgo, T. Eisenberg, S. Büttner, G. Marino, K. Rafal, P. Jansen-Dürr, K. Fröhlich, G. Kroemer, and F. Madeo, "Lifespan extension by methionine restriction requires autophagy-dependent vacuolar acidification.", in press, 2014.
- A.L. Hughes, and D.E. Gottschling, "An early age increase in vacuolar pH limits mitochondrial function and lifespan in yeast", Nature, vol. 492, pp. 261-265, 2012. http://dx.doi.org/10.1038/nature11654
- M.G. Mirisola, G. Taormina, P. Fabrizio, M. Wei, J. Hu, and V.D. Longo, "Serine- and Threonine/Valine-Dependent Activation of PDK and Tor Orthologs Converge on Sch9 to Promote Aging", PLoS Genetics, vol. 10, pp. e1004113, 2014. http://dx.doi.org/10.1371/journal.pgen.1004113
- B.S. Kristal, and Y.I. Shurubor, "Metabolomics: Opening Another Window into Aging", Science of Aging Knowledge Environment, vol. 2005, 2005. http://dx.doi.org/10.1126/sageke.2005.26.pe19
COPYRIGHT
© 2014

Metabolites in aging and autophagy by Sabrina Schroeder et al. is licensed under a Creative Commons Attribution 4.0 International License.