Research Articles:
Microbial Cell, Vol. 1, No. 11, pp. 376 - 386; doi: 10.15698/mic2014.11.173
Overexpression of the transcription factor Yap1 modifies intracellular redox conditions and enhances recombinant protein secretion
1 Department of Biotechnology, BOKU University of Natural Resources and Life Sciences Vienna, Vienna, Austria.
2 Austrian Centre of Industrial Biotechnology (ACIB), Vienna, Austria.
3 School of Bioengineering, University of Applied Sciences FH Campus Wien, Vienna, Austria.
4 Department of Chemistry, BOKU University of Natural Resources and Life Sciences Vienna, Vienna, Austria.
Keywords: ER, cytosol, cellular redox regulation, oxidative protein folding, glutathione, redox sensitive roGFP, Pichia pastoris.
Abbreviations:
ER - endoplasmic reticulum,
PDI - protein disulfide isomerase,
ROS - reactive oxygen species,
roGFP - redox sensitive variants of green fluorescent protein,
SOD - superoxide dismutase,
TRP - trypsinogen,
UPR - unfolded protein response.
Received originally: 18/04/2014 Received in revised form: 09/09/2014
Accepted: 17/10/2014
Published: 31/10/2014
Correspondence:
Brigitte Gasser, Department of Biotechnology, BOKU University of Natural Resources and Life Sciences Vienna, Muthgasse 18; 1190 Vienna, Austria brigitte.gasser@boku.ac.at
Conflict of interest statement: The authors declare no conflict of interest.
Please cite this article as: Marizela Delic, Alexandra B. Graf, Gunda Koellensperger, Christina Haberhauer-Troyer, Stephan Hann, Diethard Mattanovich, Brigitte Gasser (2014). Overexpression of the transcription factor Yap1 modifies intracellular redox conditions and enhances recombinant protein secretion. Microbial Cell 1(11): 376-386.
Abstract
Oxidative folding of secretory proteins in the endoplasmic reticulum (ER) is a redox active process, which also impacts the redox conditions in the cytosol. As the transcription factor Yap1 is involved in the transcriptional response to oxidative stress, we investigate its role upon the production of secretory proteins, using the yeast Pichia pastoris as model, and report a novel important role of Yap1 during oxidative protein folding. Yap1 is needed for the detoxification of reactive oxygen species (ROS) caused by increased oxidative protein folding. Constitutive co-overexpression of PpYAP1 leads to increased levels of secreted recombinant protein, while a lowered Yap1 function leads to accumulation of ROS and strong flocculation. Transcriptional analysis revealed that more than 150 genes were affected by overexpression of YAP1, in particular genes coding for antioxidant enzymes or involved in oxidation-reduction processes. By monitoring intracellular redox conditions within the cytosol and the ER using redox-sensitive roGFP1 variants, we could show that overexpression of YAP1 restores cellular redox conditions of protein-secreting P. pastoris by reoxidizing the cytosolic redox state to the levels of the wild type. These alterations are also reflected by increased levels of oxidized intracellular glutathione (GSSG) in the YAP1 co-overexpressing strain. Taken together, these data indicate a strong impact of intracellular redox balance on the secretion of (recombinant) proteins without affecting protein folding per se. Re-establishing suitable redox conditions by tuning the antioxidant capacity of the cell reduces metabolic load and cell stress caused by high oxidative protein folding load, thereby increasing the secretion capacity.
INTRODUCTION
Cells have evolved potent antioxidant mechanisms in order to circumvent oxidative stress and oxidative damage of cellular components when reactive oxygen species (ROS) occur during physiological conditions. Apart from the mitochondrial respiratory chain and the beta-oxidation of fatty acids occurring in the peroxisomes, also oxidative protein folding of secretory proteins within the endoplasmic reticulum (ER) was reported to contribute to the formation of ROS (reviewed by [1][2][3][4]).
–
Formation of disulfide bonds is attained through the oxidative protein folding machinery in the ER, using glutathione as redox buffer. In order to enable oxidative protein folding, the ER is specially equipped with dedicated enzymes such as oxidoreductases and chaperones and has a more oxidizing environment than the cytosol (for recent reviews see e.g. [5][6][7]). During de novo formation of disulfide bonds, protein disulfide isomerase (PDI) introduces disulfide bonds into nascent client proteins, which causes reduction of PDI. FAD-dependent ER oxidoreductin Ero1 is responsible for the re-oxidation of PDI [8]. Ero1 uses molecular oxygen as terminal electron acceptor, thereby generating stoichiometric amounts of hydrogen peroxide H2O2 [9][10]. It has been suggested that glutathione acts as a redox buffer against ER-derived oxidative stress [11][12].
–
Once generated, ROS can damage nucleic acids and lead to oxidation of proteins and peroxidation of lipids [13]. As a consequence, the expression of a set of proteins that eliminate ROS is induced [14], which is called “the oxidative stress response”. Most organisms have evolved a combination of mechanisms for maintaining cellular redox balance, involving both ROS detoxifying enzymes with very high catalytic activity (such as superoxide dismutases (SODs), catalases and glutathione peroxidases), redox-regulating enzymes (e.g., thioredoxin, glutaredoxins, peroxiredoxins) as well as non-enzymatic compounds such as glutathione [3]. The oxidative stress response is a complex regulatory circuit controlled by the interplay of different transcription factors, which are often subject to redox induced structural changes [15][4]. Among them, the yeast AP-1 transcription factor Yap1 plays a major role in the regulation of the transcriptional response to oxidative stress [16].
–
Upon treatment of the yeast Saccharomyces cerevisiae with hydrogen peroxide, at least 115 genes are overexpressed and 52 repressed as a consequence of oxidative stress in the cell [17], the majority of which is regulated by Yap1p [18]. Yap1p-mediated regulatory pathways are activated by redox-sensitive cysteine residues that serve as cellular sensors for changes in the intracellular redox balance (reviewed e.g. by [3] and [12]. Through the formation of different intramolecular disulfide bonds, Yap1 undergoes different conformational changes upon exposure to different oxidants (such as hydrogen peroxide or the superoxide generating oxidant menadione), which result in nuclear localization of Yap1 by masking the leucine-rich nuclear export signal [19]. Microarray data of a YAP1 overexpressing S. cerevisiae strain revealed that oxidoreductases formed a remarkable fraction of the regulated genes. This group was supposed to have a protective function upon oxidative stress [20]. Another important function of Yap1 and its target genes is to balance cytosolic redox homeostasis [21].
–
Using the yeast Pichia pastoris as a model, we have recently shown that protein folding stress within the ER has a strong impact on the redox state of the cytosol, leading to a significant reduction of this compartment [22]. By applying redox sensitive variants of green fluorescent protein (roGFP) to measure in vivo glutathione redox conditions in ER and cytosol during oxidative protein folding, we observed a significant reduction of the redox state of the cytosol when ER resident proteins such as Ero1, Pdi1, or recombinant secretory proteins were overexpressed.
–
Cellular redox imbalance is stressful for the cells, and does not only lead to reduced productivity in biotechnological production of recombinant proteins, but is also associated with the development of many aging-related human diseases including diabetes mellitus, atherosclerosis, and neurodegenerative diseases such as Alzheimer’s, amyotrophic lateral sclerosis (ALS) and Parkinson’s (reviewed e.g. by [23]). Therefore we aimed at restoring cytosolic redox ratios during conditions of increased oxidative protein folding in the ER.
–
Yano et al. recently identified the P. pastoris Yap1 homolog and reported the involvement of the transcription factor in the detoxification of formaldehyde and ROS in cells grown on methanol as carbon and energy source [24][25]. Here we investigated the role of Yap1 during the production of recombinant secretory proteins in glucose based growth conditions in P. pastoris, and report a novel role of Yap1 during ER resident oxidative protein folding.
RESULTS AND DISCUSSION
Downregulation of the transcription factor Yap1 leads to accumulation of reactive oxygen species (ROS), whereas Yap1 overproduction has a positive influence on protein secretion
In our previous studies we reported that increased oxidative protein folding within the ER of P. pastoris led to changes in the redox state of the cytosol, independent of UPR activation [22]. Oxidative protein folding has been implicated with the generation of stoichiometric amounts of H2O2 during regeneration of reduced Pdi1 by Ero1 [26], correspondingly, we detected ROS in strains with increased levels of Pdi1. However, we did not detect ROS accumulation upon increased folding load due to overexpression of secretory model proteins unless ER stress and UPR activation occurred [22]. Therefore we assumed that the cellular ROS scavenging system is effective to degrade ROS under physiological levels of oxidative folding, but is overwhelmed upon severe ER stress, thus leading to ROS accumulation. As Yap1 could be involved in the detoxification of ROS generated during oxidative folding of proteins in the ER, we examined the role of this transcription factor during the production of the secretory model protein trypsinogen (TRP) in more detail. In order to observe the specific effects of oxidative protein folding, we used a TRP secreting strain which is not induced for UPR [22].
–
In this strain background (trp), we investigated the influence of lowered (trp∆yap1) and increased constitutive (trpYAP1) expression levels of YAP1. Increased constitutive expression of YAP1 was obtained by integrating an additional copy of YAP1 under the control of the strong glycolytic glyceraldehyde-3-phosphate dehydrogenase PGAP promoter into the P. pastoris genome [27], while down-regulation of the YAP1 expression level was achieved through the exchange of the native promoter of the respective gene with the serine repressible promoter PSER1 [28]. Repression of this promoter of the serine biosynthesis gene SER1 was obtained through repeated addition of 10 mM serine to the synthetic M2 medium during the cultivation. We decided to go for a conditional yap1 repression rather than a total gene knock out in order to be more flexible and prevent potential growth impairment as was reported for the yap1 knock-out during growth on methanol [24][25]. We did not analyse YAP1 down-regulation in the wild type background, because so far no detectable phenotypic differences were reported for yap1 mutants during normal non-stressed growth conditions ([25] for P. pastoris, [29] for S. cerevisiae).
–
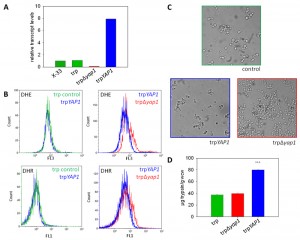
Transcript level determination with quantitative real time PCR showed clearly that the trp∆yap1 strain expressed less than 10% of YAP1 mRNA compared to P. pastoris wild type X-33 or the parental trp strain (all grown in repressing conditions of PSER1), whereas the trpYAP1 strain exhibited significantly higher YAP1 mRNA levels (approx. 8-fold higher levels as compared to X-33 and the parental trp strain) (Figure 1A).
Next, we determined the levels of accumulated ROS in the Yap1 deregulated strains compared to their parental strain. DHR and DHE were used for detection of H2O2 and superoxide anion as described previously [22]. As reported in [22], the non-UPR induced single copy trp strain did not accumulate significant levels of ROS compared to the wild type X-33 (data not shown). ROS analysis with flow cytometry revealed that trp∆yap1 indeed accumulated significantly higher levels of both ROS species compared to the non-engineered control strain (Figure 1B), indicating its essential function in detoxification of accumulated ROS. Accumulation of ROS in the YAP1 overexpressing strain trpYAP1 was indistinguishable from the non-engineered single copy trp strain. Moreover, we observed a strong phenotype of the Yap1-depleted strain trp∆yap1, which showed intensive flocculation in liquid cultures (Figure 1C). As no phenotype has been reported upon yap1 deletion previously, we hypothesize that flocculation is a response triggered by the enhanced level of oxidative protein folding.
–
Oxidative stress response has been implicated with recombinant protein secretion stress in S. cerevisiae quite recently, using a transcriptomics based approach. Although Yap1 itself did not show up within the differentially regulated genes, the Reporter Transcription Factors algorithm [30] identified Yap1 as significant based on down-regulation of Yap1 target genes in a S. cerevisiae strain secreting α-amylase, a difficult to express recombinant protein [31]. On the contrary, during our transcriptomics analyses of different recombinant protein secreting strains of P. pastoris, we did not see any significant regulation of putative Yap1 target genes involved in oxidative stress defense ([32][33] and Stadlmayr et al. unpublished data). However, a significant downregulation of YAP1, and several of its target genes such as AHP1, CTA1, GLR1, GRX3, GSH1, GSH2, SOD2, and TRX1 (among other genes) was observed in a HAC1 overexpression P. pastoris strain (except for SOD1 which was up-regulated; data from [34]) correlating with ROS accumulation in this strain with constitutive UPR induction [22].
–
Analysis of trypsinogen secretion levels in the strains deregulated for Yap1 transcript levels substantiate a role of Yap1 in the secretion process. YAP1 overexpression enhanced the level of secreted trypsinogen in the single copy trp strain more than 2-fold (Figure 1D). Notably, only the amount of correctly folded active protein is measured by the enzymatic TAME assay [35]. No influence on biomass specific trypsinogen yields were observed in the conditional yap1 knock-down strain (Figure 1D). Quantitative real time PCR analysis showed that transcript levels of genes involved in oxidative protein folding, such as ERO1 and PDI1, which have been shown to enhance trypsinogen secretion previously [22], remained unchanged in the YAP1 overexpressing strain (data not shown), ruling out one possible explanation for the enhanced secretion phenotype. Additionally, we confirmed the positive impact of YAP1 overexpression on protein secretion also in a high level secreting and UPR-induced P. pastoris strain (containing multiple copies of the pTRP expression cassette), leading on average to 1.58-fold higher secretion yields.
–
Genes involved in the antioxidant response are significantly up-regulated in the YAP1-overexpressing strain trpYAP1
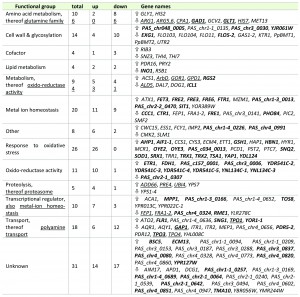
In order to elucidate the cellular mechanisms behind the improved secretion of a recombinant protein through constitutive overexpression of YAP1, transcriptional analysis of this strain and its parental strain were performed during exponential growth of the cells using DNA microarrays [34]. The yap1 knock-down strain was excluded at this point, as its strong flocculation phenotype hampered quantitative sampling. This technical limitation was not an issue as our aim was to investigate the transcriptional regulation responsible for the increased secretion phenotype, and not to elucidate the Yap1 regulon upon oxidative stress in P. pastoris. Microarray analysis revealed that the constitutive overexpression of YAP1 (without any external oxidative stimulus) exerted significant regulation of 170 genes (cutoff criteria: adjusted P-value < 0.05), 98 thereof being induced, and 72 being downregulated (Table 1). Out of these genes, 146 (more than 85%) had at least one YAP binding site (ARE) in their promoter regions (determined using Matinspector, Genomatix, see Additional File 1). Basal regulation through YAP1 overexpression without stressors has also been reported for S. cerevisiae [20][36]. Most probably, the transcriptional response without external stressor is due to overload of Crm1, the protein responsible for nuclear export of Yap1, and consequently increased levels of Yap1 localized to the nucleus (as suggested by [36]). Localization was analysed using Yap1, which was C-terminally fused to sGFP and found to be distributed all over the cell including the nucleus (data not shown).
GO Term Finder (P-value cut-off < 0.02) was used to identify significantly regulated biological processes, molecular functions and cellular components (Table 1 and Additional File 1). Among the upregulated genes, GO processes “Response to oxidative stress”, “Response to chemical stimulus”, “Metal ion transport” and “Cellular homeostasis” were the most significantly influenced biological functions, while “glutamine family amino acid metabolism”, “manganese ion transport” and “flocculation” are significantly down-regulated functions.
–
Regulation of genes coding for multidrug transporter has been reported to be dependent on Yap1 [37][38][39], but has not been shown without externally applied stress previously. Transcriptional regulation of genes involved in cellular iron and copper homeostasis might look contradictory at first, as there is no difference in the external ion concentrations, but are clearly related to the lowered levels of the GATA type repressor of the iron regulon Fep1 [40] and the repressor Fra1-2 in the YAP1 overexpressing strain. Seemingly, tight control of the uptake of these free redox active metal ions is a part of the antioxidant response. The down-regulation of the several cell wall associated genes including FLO103, FLO104, FLO11, FLO5-2 upon YAP1 overexpression may explain the strong flocculation phenotype of the yap1 knock-down strain. Down-regulation of the flocculin genes might be due to the slight up-regulation of the gene encoding Nrg1 transcriptional repressor, which seems to be a key regulator of flocculation-related genes in P. pastoris (own unpublished results).
–
Contrary to previous regulation patterns observed in S. cerevisiae upon overexpression of Yap1 or oxidative stimulus [17][20], we do not see chaperones or other folding related genes among the regulated genes in our analysis, thus ruling out their impact on the improved secretion phenotype.
–
Among the regulated genes, increased expression of genes coding for antioxidant enzymes, e.g., superoxide dismutase and peroxiredoxins, was predominant (Table 1). Strong up-regulation of the majority of genes involved in cytosolic ROS detoxification such as superoxide dismutase SOD1, a second cytosolic Cu/Zn SOD isoenzyme encoded by PAS_c034_0013, SOD co-chaperone CCS1, peroxiredoxin AHP1, sulfoxiredoxin SRX1 which is needed to reduce thioredoxin peroxidase encoded by TSA1, cytoplasmic thioredoxin peroxidase TRR1 which keeps the thioredoxin system (TRX1, TRX2) in the reduced state as well as glutathione biosynthesis enzymes gamma glutamylcysteine synthetase GSH1, cystathionine gamma-lyase CYS3 and omega glutathione transferase ECM4 was observed, indicating that they might be correlated to enhanced secretion.
–
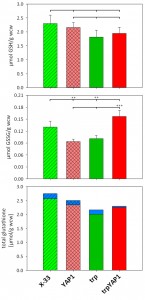
Comparison of total intra- and extracellular glutathione (GSH) and glutathione disulfide (GSSG) levels show no significant differences, but an alteration in the GSH/GSSG ratio
As glutathione biosynthesis was among the up-regulated cellular processes in trpYAP1 (GSH1, CYS3), we investigated the levels of GSH and GSSG in the YAP1 overexpressing strain and its parental control. No significant changes were determined for the concentration of reduced glutathione (Figure 2A). As shown previously [23], the strain secreting trypsinogen had a lower intracellular level of GSSG compared to the wild type strain X-33. Upon YAP1 co-overexpression, GSSG levels in the trp strain are increased more than 50% and are even slightly higher than in X-33. This effect is specific for YAP1 overexpression in the protein secreting strain, as GSSG levels were not increased in the wild type background (Figure 2B). On the contrary, GSSG levels are lower when overexpressing YAP1 in the wild type background. Total glutathione content is slightly but not significantly lower in the engineered strains (Figure 2C). It seems, that enhanced Yap1 availability is able to restore the physiological redox conditions in the secreting strain to the wild type state. Again, GSH excretion does not seem to play a significant role, as extracellular GSH levels were almost identical in all strains (Figure 2C), while extracellular GSSG concentrations are below the limit of quantification (~0.007 µmol/g wet cell weight).
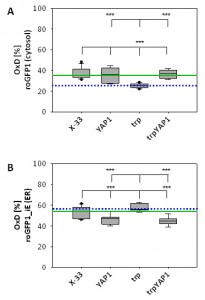
Overexpression of the transcription factor Yap1 causes a slight but significant reduction of the ER redox state, but restores cytosolic redox state in the trypsinogen secreting strain to the wild type level
As YAP1-overexpression seems to have a strong impact on intracellular redox conditions, we determined compartment-specific redox ratios in the cytosol and the ER using redox-sensitive GFP variants [27][41]. We have already demonstrated that folding of proteins in the ER does not affect the redox environment of the ER, but leads to reduction of the redox state of the cytosol [22]. Overexpression of YAP1 in the strain producing the recombinant secretory protein reoxidizes the redox state of the cytosol to the level of the wild type strain (Figure 3A), which is also reflected by the increased levels of oxidized GSSG in the trpYAP1 strain. Again, the effect is specific for the recombinant protein secreting strain, which displayed the lower cytosolic redox state. Interestingly, YAP1 overexpression had also an effect on the ER redox state, and caused a slight but significant reduction of the ER through a yet unidentified mechanism (Figure 3B).
Conclusions
Oxidative protein folding in the ER generates stoichiometric amounts of ROS, in particular H2O2. At present, it is not clear whether these ROS stay within the ER or diffuse to the cytosol, but we could clearly show that Yap1 is involved in physiological detoxification of ROS formed upon oxidative folding in the ER. Yap1 is required to activate the antioxidant enzymes, which are quenching ROS. Cells with significantly lowered Yap1 levels react to increased secretory folding load with accumulation of ROS (e.g., hydrogen peroxide and superoxide anions) and strong flocculation. On the other hand, enhanced levels of Yap1 seem to create a more convenient environment for folding and secretion of (recombinant) proteins.
–
It should be noted that protein folding is not a stoichiometric process, but usually requires several rounds of folding per molecule. It has been reported previously, that per each disulfide bond formed one molecule of hydrogen peroxide is generated [9]. Additionally, folding also requires ATP, as correct folding of nascent peptides by chaperones occurs along multiple ATP-driven cycles of substrate binding and release (recently reviewed e.g. by [42]). Thus, increased mitochondrial activity to generate sufficient amounts of ATP might also contribute to ROS formation [43]. At present we cannot distinguish between ER and mitochondria derived ROS. Based on the amount of secreted trypsinogen after 48 h in our study, at least 106 molecules of trypsinogen were processed per cell in the trp strain, with each trypsinogen having six disulfide bonds to be correctly formed. Nevertheless, we did not observe ROS accumulation in this strain compared to the non-expressing wild type strain X-33, indicating that the cell’s antioxidant response can cope with this amount of secretory proteins. However, we did see ROS accumulation in strains with higher oxidative protein folding and secretion levels [22], indicating that the additional amount of secretory load overwhelms the antioxidant capacity of the cells.
–
We have previously shown that increased oxidative folding of proteins in the ER has a strong effect on the redox environment of the cytosol, leading to more reducing conditions [22]. This cellular redox imbalance is stressful for the cells. Here we demonstrate that overexpression of YAP1 in such a strain reverses the effect on the redox status within these two compartments. It causes re-oxidation of the cytosol to the level of the wild type strain, while slightly reducing the redox state of the ER. Constitutive co-overexpression of YAP1 also leads to increased secretion levels of a recombinant protein. These alterations are also reflected by the levels of reduced and oxidized intracellular glutathione, whereby a significantly higher GSSG amount in the YAP1 co-overexpressing strain compared to its parental strain was measured. Total glutathione levels were thereby not remarkably affected among the analysed strains. Thereby, the degree of oxidation (OxD = 2*[GSSG] / [tGSH]) of the whole cell was shifted from 10% in the wild type and the trp strain to 14% in the trp_YAP1 co-overexpressing strain. Taken together, these data indicate a strong impact of intracellular redox balance on the secretion of (recombinant) proteins without affecting protein folding per se.
–
This hypothesis is further supported by microarray data of the respective P. pastoris strains, as genes involved in the antioxidant response were the predominant group of genes regulated by overexpressed YAP1, whereas no effect on the transcription of folding related genes was observed.
–
With these findings we conclude that the re-establishment of suitable redox conditions by tuning the antioxidant capacity of the cell reduces cell stress caused by recombinant protein production, and thereby increases the secretion capacity of P. pastoris. Moreover, these findings are thought to have implications not only for improving biotechnological production processes, but also impact our understanding of the development of many aging related diseases, as both cases share the underlying molecular processes, which connect cellular redox conditions with folding related stresses in the ER.
MATERIALS AND METHODS
Strains and vectors
The single copy trypsinogen expressing strain (trp) was described in [22]. Briefly, this strain contains a single expression cassette of porcine trypsinogen under control of the PGAP (glyceraldehyde-3-phosphate-dehydrogenase) promoter using the S. cerevisiae alfa-mating factor pre-pro-leader for secretion and has been confirmed not to activate the UPR by qPCR previously. For overexpression of P. pastoris YAP1, the respective gene (PAS_chr4_0601) was amplified from X-33 genomic DNA, and cloned into a pPuzzle vector [44] under control of the PGAP promoter. The Zeocin resistance marker was flanked by loxP sites. The vector was integrated into the native YAP1 locus of the P. pastoris genome after linearization in the respective sequence. After transformation by electroporation, positive transformants were selected on YPD plates with Zeocin.
–
For conditional downregulation of YAP1, the native PYAP1 promoter was exchanged for the serinerepressible PSER promoter using the strategy described in [28], resulting the trp∆yap1 strain. The used primers are summarized in Table 2.
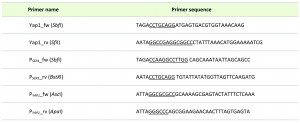 | TABLE 2. Primers used for generation of YAP1 overexpression and knock down. |
Vectors containing redox-sensitive GFP variants roGFP1 and roGFP1_iE for targeting the cytosol and the ER were described in [27] and were used for the transformation of the strains mentioned above.
–
For the localization of overproduced Yap1, superfolder GFP (sGFP) was fused to the C-terminus of the YAP1 gene, and expressed under control of PGAP. These strains were then analysed by Western blot for expression and fluorescence microscopy for intracellular localization.
–
High level trypsinogen secreting strains were generated by overexpressing pTRP under control of PGAP and selecting for the highest expressing P. pastoris strains containing multiple copies of the expression cassette on increased antibiotics (Zeocin) concentrations. For overexpression of P. pastoris YAP1 in these strains, the Zeocin resistance marker in the pPuzzle vector was exchanged for the KanMX cassette. The vector was integrated into the native YAP1 locus of the P. pastoris genome after linearization in the respective sequence. After transformation by electroporation, positive transformants were selected on YPD plates with G418.
–
Production of porcine trypsinogen
Screening media contained per liter: 10 g pea peptone, 10 g yeast extract, 10.2 g (NH)2HPO4, 1.24 g KCl, 910 µl 1 M CaCl2 solution and 1 ml biotin stock solution (0.2 g/L). Pre-culture was performed in 5 ml YPD for all strains. For the main culture, 10 clones per construct were inoculated into 10 ml of screening medium in 100 ml shake flasks (at an OD600 = 0.1) and incubated at 28°C with 170 rpm (rotations per minute) for 48 h.
–
The amount of secreted trypsinogen was determined in the supernatant by measuring the activity of enterokinase activated trypsin according to the protocol established by[45]. The sample buffer was exchanged to 1 mM HCl over PD-10 columns (Amersham Biosciences). Then, 50 µl of sample was incubated with 5 µg bovine enterokinase (Sigma) in 50 mM Tris/HCl buffer, pH 8.6, containing 50 mM CaCl2, to activate trypsinogen, and finally trypsin activity was determined with the TAME assay. Therefore, aliquots of the samples were added to 40 mM Tris/HCl buffer pH 8.1 buffer containing 10 mM CaCl2 and 1 mM p-toluene sulfonyl-L-arginine ethylester-HCl (TAME), and the increase of absorbance at 247 nm was followed. Trypsinogen concentration is calculated using the extinction coefficient ϵ = 0.0101 E247 cm-1 min-1.
–
Staining of reactive oxygen species
The fluorescent probes DHE (dihydroethidium) and DHR (dihydrorhodamine 123) were used to determine levels of ROS as described in [22]. Briefly, cells of an OD600 = 0.4 were harvested by centrifugation and resuspended in 2 ml PBS. DHE and DHR were added to cells at a final concentration of 10 μg/ml and incubated at 30°C for 30 min. Flow cytometry analysis was performed using a FACS Canto with the settings: DHR green filter (525 – 550 nm), DHE red filter (600 – 650 nm).
–
Analysis of transcriptional regulation using DNA microarrays
The strains trp and trpYAP1 were cultivated in screening medium in 4 biological replicates each. 10 ml of main culture were inoculated from an overnight pre-culture with an OD600 = 4. After 6 h (exponential growth phase), samples were taken and fixed in phenol/ethanol, and stored at -80°C until total RNA extraction. Total RNA extraction, two-color labelling in a dye swap manner for each sample and cRNA hybridization to the P. pastoris specific microarrays were performed as in [34]. Samples were hybridized to the microarrays described in Graf et al. ([34]; 8x15K custom arrays, AMAD-ID 018045, Agilent) and to a novel in house designed P. pastoris specific oligonucleotide array, which was based on the improved annotation of P. pastoris strain GS115 (8 x 15 K custom arrays, AMAD-ID 034821, Agilent, [46]). Raw data normalization was done using locally weighted MA-scatterplot smoothing (LOESS) followed by a between array ‘Aquantile` normalization, both available within the limma package of R [47]. P-values were corrected for multiple testing using Benjamini & Yekutieli method [48]. Features were defined as differentially expressed if they had a P-value < 0.05. For the identification of stronger regulatory effects an additional cut-off for the fold change (FC) of ± 1.5 fold was applied. A high degree of correlation of significantly regulated genes between the two microarrays was determined (R2 = 0.902), thus the average FC of both microarrays was used.
–
Significantly regulated P. pastoris genes with a functional annotation were categorized using the “generic gene ontology (GO) term mapper“ (http://go.princeton.edu/cgi-bin/GOTermMapper).
–
Determination of transcript levels by quantitative real time PCR
RNA isolation, cDNA synthesis and measurement of mRNA transcript levels using real time PCR was performed as described in [44] using the primers given in [22]. The reference gene for normalization was actin (ACT1), each gene transcript was correlated to ACT1 as internal control. The wild type strain X-33 served as reference strain for relative transcript level determination using the delta-delta Ct method.
–
Cultivation conditions for redox and glutathione measurements
M2 minimal medium contained per liter: 20 g of glucose, 20 g of citric acid, 3.15 g of (NH4)2HPO4, 0.03 g of CaCl2 • 2H2O, 0.8 g of KCl, 0.5 g of MgSO4 • 7H2O, 2 ml of biotin (0.2 g/L), 1.5 ml of trace salts stock solution. The pH was set to 5.0 with 5 M KOH solution. Trace salts stock solution contained per liter: 6.0 g of CuSO4 • 5H2O, 0.08 g of NaI, 3.0 g of MnSO4 •H2O, 0.2 g of Na2MoO4 • 2H2O, 0.02 g of H3BO3, 0.5 g of CoCl2, 20.0 g of ZnCl2, 5.0 g of FeSO4 • 7H2O, and 5.0 ml of H2SO4 (95-98% w/w).
–
Pre-culture was performed in 5 ml M2 medium. 10 ml M2 medium in 100 ml shake flasks were inoculated at an OD600 = 0.5, and were incubated at 28°C with 170 rpm (rotations per minute) for 24 h. For each strain, 12 individual clones were analysed.
–
Determination of redox ratios using roGFP
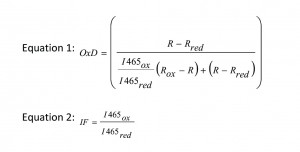
Redox statesof all strains were measured during the exponential growth phase. Briefly, 840 µl culture were mixed with 60 µl 1.5 M K-MOPS buffer (pH 7.0). Total oxidation or reduction of the roGFPs was achieved by addition of 200 µl 6.3 mM 4,4′-dipyridyl disulfide (4-DPS) or 200 µl 1 M dithiothreitol (DTT), respectively, and was compared to the untreated culture where 200 µl of PBS were added as control (Ostergaard et al. 2004, [49]). The fluorescence of the cells was detected in 96-well plates (FluoroNunc, Nunc) on a fluorescence photometer (Infinite M200 Tecan plate reader), where it was possible to measure the excitation of two wavelengths, 395 and 465 nm, corresponding to the oxidized and the reduced form of the protein. Each sample was measured four times [27]. The degree of oxidation (OxD) of the cytosol and the ER was calculated according to equation 1. The quotient of fluorescence intensities (equation 2) is the so called ‘instrument factor IF’ and corrects for variations in the signal strength from the instrument. R is the experimentally determined fluorescence ratio of the intensities for the two wavelengths 395 and 465 nm [50]. Rox and Rred stand for the ratios of fully oxidized or fully reduced roGFP, respectively, while R is the ratio of the untreated sample.
 |
Determination of glutathione concentrations by LC/MS-MS
Three pellets of 1 ml of exponential culture were collected by centrifugation at 3000 rpm for 5 min at 4°C. The cells were resuspended in 1 ml ice cold 0.1 M H3PO4, spiked with labeled glutathione-glycine-13C2,15N (Sigma Aldrich) and GSSG- glutathione-glycine-13C2,15N (synthesized in house)and heated to 75°C for 3 min to extract GSH and GSSG. The cell extracts were measured by LC/MS-MS (Agilent Ion Trap) using hydrophilic interaction chromatography (HILIC) and acetonitrile/water as eluent [51].
–
Statistical analyses
Secreted trypsinogen labels and glutathione concentrations are expressed as mean ± standard error of the mean. Statistically significant differences between the strains were determined using Student’s t-test. All strains were compared to the wild type strain X-33 or the single copy trypsinogen secreting strain trp. The P-values are marked with *** for P< 0.01 and ** for P< 0.05.
References
- Y. Shimizu, and L.M. Hendershot, "Oxidative Folding: Cellular Strategies for Dealing with the Resultant Equimolar Production of Reactive Oxygen Species", Antioxidants & Redox Signaling, vol. 11, pp. 2317-2331, 2009. http://dx.doi.org/10.1089/ARS.2009.2501
- J.D. Malhotra, and R.J. Kaufman, "Endoplasmic Reticulum Stress and Oxidative Stress: A Vicious Cycle or a Double-Edged Sword?", Antioxidants & Redox Signaling, vol. 9, pp. 2277-2294, 2007. http://dx.doi.org/10.1089/ars.2007.1782
- E. Herrero, J. Ros, G. Bellí, and E. Cabiscol, "Redox control and oxidative stress in yeast cells", Biochimica et Biophysica Acta (BBA) - General Subjects, vol. 1780, pp. 1217-1235, 2008. http://dx.doi.org/10.1016/j.bbagen.2007.12.004
- K.A. Morano, C.M. Grant, and W.S. Moye-Rowley, "The response to heat shock and oxidative stress in Saccharomyces cerevisiae.", Genetics, 2011. http://www.ncbi.nlm.nih.gov/pubmed/22209905
- �. Margittai, and R. Sitia, "Oxidative Protein Folding in the Secretory Pathway and Redox Signaling Across Compartments and Cells", Traffic, vol. 12, pp. 1-8, 2010. http://dx.doi.org/10.1111/j.1600-0854.2010.01108.x
- K. Araki, and K. Nagata, "Protein Folding and Quality Control in the ER", Cold Spring Harbor Perspectives in Biology, vol. 3, pp. a007526-a007526, 2011. http://dx.doi.org/10.1101/cshperspect.a007526
- A. Görlach, P. Klappa, and D.T. Kietzmann, "The Endoplasmic Reticulum: Folding, Calcium Homeostasis, Signaling, and Redox Control", Antioxidants & Redox Signaling, vol. 8, pp. 1391-1418, 2006. http://dx.doi.org/10.1089/ars.2006.8.1391
- A.R. Frand, and C.A. Kaiser, "Ero1p Oxidizes Protein Disulfide Isomerase in a Pathway for Disulfide Bond Formation in the Endoplasmic Reticulum", Molecular Cell, vol. 4, pp. 469-477, 1999. http://dx.doi.org/10.1016/s1097-2765(00)80198-7
- E. Gross, C.S. Sevier, N. Heldman, E. Vitu, M. Bentzur, C.A. Kaiser, C. Thorpe, and D. Fass, "Generating disulfides enzymatically: Reaction products and electron acceptors of the endoplasmic reticulum thiol oxidase Ero1p", Proceedings of the National Academy of Sciences, vol. 103, pp. 299-304, 2006. http://dx.doi.org/10.1073/pnas.0506448103
- B.P. Tu, and J.S. Weissman, "The FAD- and O2-Dependent Reaction Cycle of Ero1-Mediated Oxidative Protein Folding in the Endoplasmic Reticulum", Molecular Cell, vol. 10, pp. 983-994, 2002. http://dx.doi.org/10.1016/s1097-2765(02)00696-2
- S. Chakravarthi, C.E. Jessop, and N.J. Bulleid, "The role of glutathione in disulphide bond formation and endoplasmic‐reticulum‐generated oxidative stress", EMBO reports, vol. 7, pp. 271-275, 2006. http://dx.doi.org/10.1038/sj.embor.7400645
- M.B. Toledano, A. Delaunay-Moisan, C.E. Outten, and A. Igbaria, "Functions and Cellular Compartmentation of the Thioredoxin and Glutathione Pathways in Yeast", Antioxidants & Redox Signaling, vol. 18, pp. 1699-1711, 2013. http://dx.doi.org/10.1089/ars.2012.5033
- M.D. Temple, G.G. Perrone, and I.W. Dawes, "Complex cellular responses to reactive oxygen species", Trends in Cell Biology, vol. 15, pp. 319-326, 2005. http://dx.doi.org/10.1016/j.tcb.2005.04.003
- K. Linke, and U. Jakob, "Not Every Disulfide Lasts Forever: Disulfide Bond Formation as a Redox Switch", Antioxidants & Redox Signaling, vol. 5, pp. 425-434, 2003. http://dx.doi.org/10.1089/152308603768295168
- N. Brandes, S. Schmitt, and U. Jakob, "Thiol-Based Redox Switches in Eukaryotic Proteins", Antioxidants & Redox Signaling, vol. 11, pp. 997-1014, 2009. http://dx.doi.org/10.1089/ARS.2008.2285
- K.D. Harshman, W. Moye-Rowley, and C.S. Parker, "Transcriptional activation by the SV40 AP-1 recognition element in yeast is mediated by a factor similar to AP-1 that is distinct from GCN4", Cell, vol. 53, pp. 321-330, 1988. http://dx.doi.org/10.1016/0092-8674(88)90393-5
- C. Godon, G. Lagniel, J. Lee, J. Buhler, S. Kieffer, M. Perrot, H. Boucherie, M.B. Toledano, and J. Labarre, "The H2O2 Stimulon in Saccharomyces cerevisiae", Journal of Biological Chemistry, vol. 273, pp. 22480-22489, 1998. http://dx.doi.org/10.1074/jbc.273.35.22480
- J. Lee, C. Godon, G. Lagniel, D. Spector, J. Garin, J. Labarre, and M.B. Toledano, "Yap1 and Skn7 Control Two Specialized Oxidative Stress Response Regulons in Yeast", Journal of Biological Chemistry, vol. 274, pp. 16040-16046, 1999. http://dx.doi.org/10.1074/jbc.274.23.16040
- S. Kuge, T. Toda, N. Iizuka, and A. Nomoto, "Crm1 (XpoI) dependent nuclear export of the budding yeast transcription factor yAP‐1 is sensitive to oxidative stress", Genes to Cells, vol. 3, pp. 521-532, 1998. http://dx.doi.org/10.1046/j.1365-2443.1998.00209.x
- J.L. DeRisi, V.R. Iyer, and P.O. Brown, "Exploring the Metabolic and Genetic Control of Gene Expression on a Genomic Scale", Science, vol. 278, pp. 680-686, 1997. http://dx.doi.org/10.1126/science.278.5338.680
- A. Ayer, S. Fellermeier, C. Fife, S.S. Li, G. Smits, A.J. Meyer, I.W. Dawes, and G.G. Perrone, "A Genome-Wide Screen in Yeast Identifies Specific Oxidative Stress Genes Required for the Maintenance of Sub-Cellular Redox Homeostasis", PLoS ONE, vol. 7, pp. e44278, 2012. http://dx.doi.org/10.1371/journal.pone.0044278
- M. Delic, C. Rebnegger, F. Wanka, V. Puxbaum, C. Haberhauer-Troyer, S. Hann, G. Köllensperger, D. Mattanovich, and B. Gasser, "Oxidative protein folding and unfolded protein response elicit differing redox regulation in endoplasmic reticulum and cytosol of yeast", Free Radical Biology and Medicine, vol. 52, pp. 2000-2012, 2012. http://dx.doi.org/10.1016/j.freeradbiomed.2012.02.048
- H. Yoshida, "ER stress and diseases", The FEBS Journal, vol. 274, pp. 630-658, 2007. http://dx.doi.org/10.1111/j.1742-4658.2007.05639.x
- T. YANO, H. YURIMOTO, and Y. SAKAI, "Activation of the Oxidative Stress Regulator PpYap1 through Conserved Cysteine Residues during Methanol Metabolism in the YeastPichia pastoris", Bioscience, Biotechnology, and Biochemistry, vol. 73, pp. 1404-1411, 2009. http://dx.doi.org/10.1271/bbb.90109
- T. Yano, E. Takigami, H. Yurimoto, and Y. Sakai, "Yap1-Regulated Glutathione Redox System Curtails Accumulation of Formaldehyde and Reactive Oxygen Species in Methanol Metabolism of Pichia pastoris", Eukaryotic Cell, vol. 8, pp. 540-549, 2009. http://dx.doi.org/10.1128/EC.00007-09
- B.P. Tu, and J.S. Weissman, "Oxidative protein folding in eukaryotes", The Journal of Cell Biology, vol. 164, pp. 341-346, 2004. http://dx.doi.org/10.1083/jcb.200311055
- M. Delic, D. Mattanovich, and B. Gasser, "Monitoring intracellular redox conditions in the endoplasmic reticulum of living yeasts", FEMS Microbiology Letters, vol. 306, pp. 61-66, 2010. http://dx.doi.org/10.1111/j.1574-6968.2010.01935.x
- M. Delic, D. Mattanovich, and B. Gasser, "Repressible promoters – A novel tool to generate conditional mutants in Pichia pastoris", Microbial Cell Factories, vol. 12, 2013. http://dx.doi.org/10.1186/1475-2859-12-6
- J.A. Wemmie, A.L. Wu, K.D. Harshman, C.S. Parker, and W.S. Moye-Rowley, "Transcriptional activation mediated by the yeast AP-1 protein is required for normal cadmium tolerance.", The Journal of biological chemistry, 1994. http://www.ncbi.nlm.nih.gov/pubmed/8182076
- A.P. Oliveira, K.R. Patil, and J. Nielsen, "Architecture of transcriptional regulatory circuits is knitted over the topology of bio-molecular interaction networks", BMC Systems Biology, vol. 2, 2008. http://dx.doi.org/10.1186/1752-0509-2-17
- K.E. Tyo, Z. Liu, D. Petranovic, and J. Nielsen, "Imbalance of heterologous protein folding and disulfide bond formation rates yields runaway oxidative stress", BMC Biology, vol. 10, 2012. http://dx.doi.org/10.1186/1741-7007-10-16
- K. Baumann, M. Carnicer, M. Dragosits, A.B. Graf, J. Stadlmann, P. Jouhten, H. Maaheimo, B. Gasser, J. Albiol, D. Mattanovich, and P. Ferrer, "A multi-level study of recombinant Pichia pastoris in different oxygen conditions", BMC Systems Biology, vol. 4, 2010. http://dx.doi.org/10.1186/1752-0509-4-141
- M. Dragosits, J. Stadlmann, A. Graf, B. Gasser, M. Maurer, M. Sauer, D.P. Kreil, F. Altmann, and D. Mattanovich, "The response to unfolded protein is involved in osmotolerance of Pichia pastoris", BMC Genomics, vol. 11, 2010. http://dx.doi.org/10.1186/1471-2164-11-207
- A. Graf, B. Gasser, M. Dragosits, M. Sauer, G.G. Leparc, T. Tüchler, D.P. Kreil, and D. Mattanovich, "Novel insights into the unfolded protein response using Pichia pastoris specific DNA microarrays", BMC Genomics, vol. 9, 2008. http://dx.doi.org/10.1186/1471-2164-9-390
- K. Walsh, and P. Wilcox, "[3] Serine proteases", Methods in Enzymology, pp. 31-41, 1970. http://dx.doi.org/10.1016/0076-6879(70)19005-7
- I. Nisamedtinov, K. Kevvai, K. Orumets, L. Arike, I. Sarand, M. Korhola, and T. Paalme, "Metabolic changes underlying the higher accumulation of glutathione in Saccharomyces cerevisiae mutants", Applied Microbiology and Biotechnology, vol. 89, pp. 1029-1037, 2010. http://dx.doi.org/10.1007/s00253-010-2946-z
- B. Oskouian, and J.D. Saba, "YAP1 confers resistance to the fatty acid synthase inhibitor cerulenin through the transporter Flr1p in Saccharomyces cerevisiae", Molecular and General Genetics MGG, vol. 261, pp. 346-353, 1999. http://dx.doi.org/10.1007/s004380050975
- D. Nguyên, A. Alarco, and M. Raymond, "Multiple Yap1p-binding Sites Mediate Induction of the Yeast Major Facilitator FLR1 Gene in Response to Drugs, Oxidants, and Alkylating Agents", Journal of Biological Chemistry, vol. 276, pp. 1138-1145, 2001. http://dx.doi.org/10.1074/jbc.m008377200
- K. Miyahara, D. Hirata, and T. Miyakawa, "yAP-1- and yAP-2-mediated, heat shock-induced transcriptional activation of the multidrug resistance ABC transporter genes inSaccharomyces cerevisiae", Current Genetics, vol. 29, pp. 103-105, 1996. http://dx.doi.org/10.1007/bf02221572
- R. Miele, D. Barra, and M.C. Bonaccorsi di Patti, "A GATA-type transcription factor regulates expression of the high-affinity iron uptake system in the methylotrophic yeast Pichia pastoris", Archives of Biochemistry and Biophysics, vol. 465, pp. 172-179, 2007. http://dx.doi.org/10.1016/j.abb.2007.05.020
- J.R. Lohman, and S.J. Remington, "Development of a Family of Redox-Sensitive Green Fluorescent Protein Indicators for Use in Relatively Oxidizing Subcellular Environments", Biochemistry, vol. 47, pp. 8678-8688, 2008. http://dx.doi.org/10.1021/bi800498g
- T. Simmen, E.M. Lynes, K. Gesson, and G. Thomas, "Oxidative protein folding in the endoplasmic reticulum: Tight links to the mitochondria-associated membrane (MAM)", Biochimica et Biophysica Acta (BBA) - Biomembranes, vol. 1798, pp. 1465-1473, 2010. http://dx.doi.org/10.1016/j.bbamem.2010.04.009
- R.U.H. Mattoo, and P. Goloubinoff, "Molecular chaperones are nanomachines that catalytically unfold misfolded and alternatively folded proteins", Cellular and Molecular Life Sciences, vol. 71, pp. 3311-3325, 2014. http://dx.doi.org/10.1007/s00018-014-1627-y
- G. Stadlmayr, A. Mecklenbräuker, M. Rothmüller, M. Maurer, M. Sauer, D. Mattanovich, and B. Gasser, "Identification and characterisation of novel Pichia pastoris promoters for heterologous protein production", Journal of Biotechnology, vol. 150, pp. 519-529, 2010. http://dx.doi.org/10.1016/j.jbiotec.2010.09.957
- H. Hohenblum, B. Gasser, M. Maurer, N. Borth, and D. Mattanovich, "Effects of gene dosage, promoters, and substrates on unfolded protein stress of recombinant Pichia pastoris", Biotechnology and Bioengineering, vol. 85, pp. 367-375, 2003. http://dx.doi.org/10.1002/bit.10904
- R. Prielhofer, M. Maurer, J. Klein, J. Wenger, C. Kiziak, B. Gasser, and D. Mattanovich, "Induction without methanol: novel regulated promoters enable high-level expression in Pichia pastoris", Microbial Cell Factories, vol. 12, 2013. http://dx.doi.org/10.1186/1475-2859-12-5
- G.K. Smyth, "limma: Linear Models for Microarray Data", Bioinformatics and Computational Biology Solutions Using R and Bioconductor, pp. 397-420, . http://dx.doi.org/10.1007/0-387-29362-0_23
- A. Reiner, D. Yekutieli, and Y. Benjamini, "Identifying differentially expressed genes using false discovery rate controlling procedures", Bioinformatics, vol. 19, pp. 368-375, 2003. http://dx.doi.org/10.1093/bioinformatics/btf877
- H. Østergaard, C. Tachibana, and J.R. Winther, "Monitoring disulfide bond formation in the eukaryotic cytosol", The Journal of Cell Biology, vol. 166, pp. 337-345, 2004. http://dx.doi.org/10.1083/jcb.200402120
- A.J. Meyer, and T.P. Dick, "Fluorescent Protein-Based Redox Probes", Antioxidants & Redox Signaling, vol. 13, pp. 621-650, 2010. http://dx.doi.org/10.1089/ars.2009.2948
- C. Haberhauer-Troyer, M. Delic, B. Gasser, D. Mattanovich, S. Hann, and G. Koellensperger, "Accurate quantification of the redox-sensitive GSH/GSSG ratios in the yeast Pichia pastoris by HILIC–MS/MS", Analytical and Bioanalytical Chemistry, vol. 405, pp. 2031-2039, 2012. http://dx.doi.org/10.1007/s00216-012-6620-4
SUPPLEMENTAL INFORMATION
![]() Download Supplemental Information
Download Supplemental Information
ACKNOWLEDGMENTS
The authors thank Dr. Martin Dragosits, Roland Prielhofer and Caroline Kotlowski for microarray hybridization, Corinna Rebnegger and Franziska Wanka for their help with qRT-PCR during the course of their diploma theses, Christine Sanystra and Martin Nagl for generation of high producing trp strains and their analysis, Stefan Pflügl for HPLC measurements, Verena Puxbaum for performing fluorescence microscopy and also for microsome preparation and glutathione assay together with Katharina Littringer. The authors declare that they have no competing interests.
-
Parts of this work have been supported by the Federal Ministry of Science, Research and Economy (BMWFW), the Federal Ministry of Traffic, Innovation and Technology (bmvit), the Styrian Business Promotion Agency SFG, the Standortagentur Tirol and ZIT – Technology Agency of the City of Vienna through the COMET-Funding Program managed by the Austrian Research Promotion Agency FFG. EQ BOKU VIBT GmbH is acknowledged for providing LC-MS/MS instrumentation. Finally, we thank the BOKU-VIBT Imaging Center for access and expertise with fluorescence microscopy equipment.
COPYRIGHT
© 2014

Overexpression of the transcription factor Yap1 modifies intracellular redox conditions and enhances recombinant protein secretion by Marizela Delic et al. is licensed under a Creative Commons Attribution 4.0 International License.