Research Articles:
Microbial Cell, Vol. 1, No. 3, pp. 94 - 99; doi: 10.15698/mic2014.01.131
Salt stress causes cell wall damage in yeast cells lacking mitochondrial DNA
1 State Key Laboratory of Bioreactor Engineering, East China University of Science and Technology, 130 Meilong Road, Shanghai 200237, China.
2 Department of Zoology and Physiology, University of Wyoming, Laramie, WY 82071, USA.
3 State Key Laboratory of Microbial Technology, Shandong University, Jinan, China.
Keywords: cell wall damage; reactive oxygen species; SCW11; salt stress; Saccharomyces cerevisiae.
Received originally: 11/01/2014 Received in revised form: 18/02/2014
Accepted: 26/02/2014
Published: 03/03/2014
Correspondence:
Dr. Zhaojie Zhang, Department of Zoology and Physiology, University of Wyoming; Laramie, WY, 82071, USA zzhang@uwyo.edu
Conflict of interest statement: The authors declare no conflict of interest.
Please cite this article as: Qiuqiang Gao, Liang-Chun Liou, Qun Ren, Xiaoming Bao and Zhaojie Zhang (2014). Salt stress causes cell wall damages in yeast cells lacking mitochondrial DNA. Microbial Cell 1(3): 94-99.
Abstract
The yeast cell wall plays an important role in maintaining cell morphology, cell integrity and response to environmental stresses. Here, we report that salt stress causes cell wall damage in yeast cells lacking mitochondrial DNA (ρ0). Upon salt treatment, the cell wall is thickened, broken and becomes more sensitive to the cell wall-perturbing agent sodium dodecyl sulfate (SDS). Also, SCW11 mRNA levels are elevated in ρ0 cells. Deletion of SCW11 significantly decreases the sensitivity of ρ0 cells to SDS after salt treatment, while overexpression of SCW11 results in higher sensitivity. In addition, salt stress in ρ0 cells induces high levels of reactive oxygen species (ROS), which further damages the cell wall, causing cells to become more sensitive towards the cell wall-perturbing agent.
INTRODUCTION
The yeast Saccharomyces cerevisiae cell wall occupies about 15% of the total cell volume and plays important roles in maintaining cell shape, cell integrity and protection against environmental stresses [1]. The yeast cell wall forms a microfibrillar network complex composed of glucan, mannoprotein and chitin. Glucan is mainly (80 to 90%) composed of β-1,3-glucan chains with some β-1,6-linked glucan branches. Glucan represents 50-60% of the cell wall mass and its main function is to maintain cell elasticity that can adapt to different physiological states (sporulation or budding) and various stress conditions, including exposure to cell wall-perturbing agents or hypo-osmotic shock [2][3].
–
Scw11p is a cell wall protein similar to glucanases, whose main function is to break down glucans [4][5]. Whole-genome transcriptional analysis suggests that Scw11p is required for efficient cell separation following cytokinesis [6], and is also associated with cell wall metabolism [7]. Mutation of scw11 suppresses the phenotype of Dscw4/Dscw10 double mutants, or Dscw4/Dscw10/ Dbgl2 triple mutants, suggesting that Scw11p has an activity antagonistic to that of Scw4p, Scw10p and Bgl2p [8].
–
Reactive oxygen species (ROS) play a crucial role in the signal transduction pathways that regulate yeast cell death [9][10]. Several studies have shown that ROS function as inducers of yeast apoptosis [11], while others have demonstrated that ROS play a crucial role as mediators of apoptosis [12][13]. It has been shown that ROS accumulate in yeast cells after induction of apoptotic death by various stimuli, and are necessary and sufficient to induce an apoptotic phenotype in yeast [11][14][15][16].
–
Previously, we showed that yeast lacking mitochondrial DNA (ρ0) undergoes apoptosis upon salt stress and that cell death is mediated by cytochrome c [17]. In this study, we further report that salt stress causes cell wall damage (CWD) in ρ0 cells and that this damage is related to elevated levels of SCW11 and salt stress-induced ROS.
RESULTS AND DISCUSSION
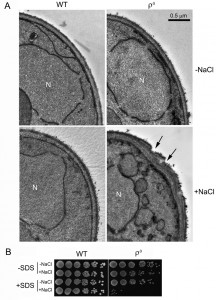
Salt stress causes cell wall damage (CWD) in ρ0 cells
We previously reported that ρ0 cells were sensitive to salt stress and consequently underwent apoptotic cell death [17]. In the current study, our TEM analysis revealed that salt stress caused an abnormal cell wall structure in ρ0 cells. When treated with NaCl, the cell wall of more than 50% of the ρ0 cells became thicker and uneven, compared to ρ0 cells without NaCl treatment or to the wild type (WT). Certain areas of the cell wall also appeared abrupt, or damaged (Fig. 1A). To further examine possible salt stress effects on the cell wall, we tested the sensitivity of ρ0 cells towards sodium dodecyl sulfate (SDS), a typical cell wall-perturbing agent, before and after salt stress. As shown in Fig. 1B, no apparent change was observed in WT cells when treated with NaCl, SDS or both. ρ0 cells, however, were hypersensitive to SDS after salt stress compared to cells treated with salt or SDS alone, further suggesting that the cell wall was damaged by the salt stress.
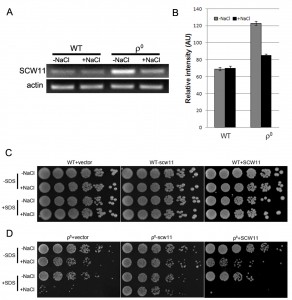
SCW11 is involved in CWD of ρ0 cells under salt stress
Scw11p is a cell wall protein with an endo-1,3-β-glucanase activity. It has been previously reported that the SCW11 gene is up-regulated in ρ0 cells [18]. However, it is still elusive how SCW11 is involved in the salt stress response in ρ0 cells. In this study, SCW11 gene expression was first examined using RT-PCR with or without salt treatment. Consistent with microarray analyses [18], we showed that in the absence of NaCl, the SCW11 expression level was significantly (P < 0.001) higher in ρ0 cells than in the WT cells. Under salt stress conditions, SCW11 expression in ρ0 cells was decreased but still significantly higher (P < 0.05) than in WT cells (Fig. 2A, 2B).
To further assess the role of SCW11 during salt stress response, SCW11 was deleted or overexpressed in both WT and ρ0 cells. As shown in Fig. 2C, deletion or overexpression of SCW11 had no apparent effect on the WT. However, deletion of SCW11 greatly enhanced the survival rate of ρ0 cells, especially when the cells were treated with both NaCl and SDS. SCW11 overexpression caused ρ0 cells to become more sensitive towards salt stress and SDS (Fig. 2D). These results suggest that, under salt stress conditions, SCW11 is involved in cell wall organization and its downregulation in ρ0 cells (Fig. 2A) is part of a protection mechanism against salt stress.
–
The yeast cell wall is a dynamic structure that can adapt to different physiological states and various stress conditions [2][3]. The endo-β-1,3-glucanase is an abundant protein in the yeast cell wall making it rigid by inserting intra-chain linkages into 1,3-β-glucan [19]. Overexpression of the endo-β-1,3-glucanase Bgl2p has been shown to lead to defects in the cell wall structure and to sensitivity towards osmotic stress [20]. Similar to this result, we show that the SCW11 gene, coding for a glucanase like-protein, is upregulated in ρ0 cells, where it leads to a higher sensitivity towards SDS and salt stress.
–
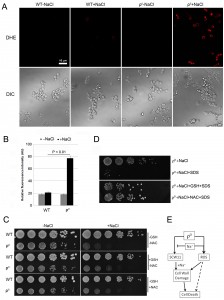
ROS is involved in CWD of ρ0 cells under salt stress
Mitochondria are the main source for ROS, which play an important role in yeast apoptosis. With the lack of mitochondrial DNA, it is unclear if ρ0 cells produce any ROS, and if ROS are involved in salt stress response of the ρ0 cells. We first examined the ROS level using dihydroethidium (DHE), a fluorescence dye that has specificity toward superoxide for detecting ROS production [16][21][22]. Little ROS was detected in wild type in the absence or presence of NaCl. In ρ0 cells, however, NaCl treatment caused a significant increase of the ROS level (Fig. 3A, 3B). Addition of antioxidant GSH or NAC only slightly improved the cell survival of ρ0 cells (Fig. 3C), suggesting that ROS may not directly be involved in salt stress response and cell death of the ρ0 cells. We further tested if addition of the antioxidant could improve the sensitivity of ρ0 cells to SDS. As shown in Fig. 3D, in the presence of GSH or NAC, ρ0 cells were much less sensitive to SDS after NaCl treatment. This result suggests that ROS likely affect the salt sensitivity of ρ0 cells, at least in part, via their damage to cell wall. Deletion or overexpression of SCW11 in ρ0 cells greatly affects the sensitivity of ρ0 cells towards SDS after the salt stress (Fig. 2D). However, DHE staining showed that deletion or overexpression of SCW11 did not affect the ROS level of the ρ0 cells (Fig. S1, compared to Figs. 3A, 3B). This result further suggests that ROS are not directly involved in the cell survival or cell death of ρ0 cells, but rather through their damage to cell wall. Deletion of SCW11 makes ρ0 cells more resistant, while SCW11 overexpression makes them more sensitive, to ROS damage.
It has been reported that ROS cause damages to DNA, proteins and lipids [23], which in turn, induce apoptotic cell death [11][24], or aging [25]. To our best knowledge, this is the first report that ROS induce potential cell wall damage in ρ0 cells. One possibility is that the increased level of Scw11 makes the cell wall more vulnerable to ROS.
–
In summary, we demonstrated that the SCW11 level was elevated in ρ0 cells and the high level of Scw11 caused ρ0 cells prone to cell wall damage by salt stress. The salt stress-induced ROS further damaged the cell wall, causing decreased viability and cell death of the ρ0 cells (Fig. 3E).
MATERIALS AND METHODS
Yeast strains and media
Yeast strains are derivatives of S. cerevisiae W303-1A (MATa, leu2-3/112 ura3-1 trp1-1 his3-11/15 ade2-1 can1-100). The scw11 deletion was introduced by PCR-mediated gene replacement [26], replacing the complete sequence of YGL028C with G418 marker. SCW11 overexpression was constructed using the BG1805 plasmid [27]. Yeast cells were routinely grown in YPD medium (1% yeast extract, 2% peptone, 2% glucose) at 30οC. For overexpression of SCW11, cells were preincubated in BG1805 selection medium (URA minus), then transferred to YPR (1% yeast extract, 2% peptone, 2% raffinose) overnight, and then to YPG (1% yeast extract, 2% peptone, 2% galactose).
–
Transmission electron microscopy (TEM)
Cells were cultured at 30οC in YPD medium to early log phase, treated with 0.6 M NaCl for 1 hr. Cells were then harvested and prepared for TEM according to [28].
–
Spot dilution growth assays
Cells were precultured in YPD liquid medium overnight. Cells were harvested by centrifugation, washed three times with ddH2O, and then resuspended in ddH2O. The cell density was normalized to 1×107 cells/ml. A five-fold serial dilution was made and 4 µl of each dilution was spotted onto appropriate YPD plates. For YPG medium growth, cells were precultured in YPR liquid overnight, and transferred to YPG liquid for 6 hr. A five-fold serial dilution was made as above, and spotted onto appropriate YPG plates. The antioxidants glutathione (GSH, 10 mM) or N-acetyl-L-cysteine (NAC, 20 mM) were added directly to the medium.
–
For the SDS sensitivity test, cells were grown in YPD liquid medium to early log phase, treated with 0.6 M NaCl (0.2 M NaCl for YPG mdium) for 1 hr, then washed with ddH2O, resuspended in 2-[4-(2-Hydroxyethyl)-1-piperazinyl] ethanesulfonic acid (HEPES) buffer (10 mM pH 7.5), treated with 0.1% SDS for 0.5 hr, then washed one time with HEPES buffer. A five-fold serial dilution was made as above, and then spotted onto the YPD or YPG plates.
–
Semi-quantitative RT-PCR
Total RNA from yeast was extracted using RNeasy Protect Mini Kit (QIAGEN, CA). The RT-PCR and the amplification procedure were performed as in [29]. Yeast actin gene (ACT1) was used as control, and the primers are 5’-TGTCACCAACTGGGACGATA-3’ and 5’-AACCAGCGTAAATTGGAACG-3’. Primers used for SCW11 amplification are 5’-CCCCAACTGTCGAATTCCTA-3’ and 5’-AAAGTAGAGGTTGGCTGCGA-3’. Quantitative analysis was conducted using ImageJ software.
–
Detection of reactive oxygen species (ROS)
The detection of ROS was performed according to [15]. Briefly, cells were grown to early log phase, treated with 0.8 M NaCl for 15 min, then washed with PBS for 3 times. Cells were stained with dihydroethidium (5 µM) (Sigma Chemical Co.) for 10 min in the dark, viewed with a Zeiss 710 laser scanning confocal microscope with excitation at 514 nm (Jena, Germany).
–
Statistical Analysis of Data
A two-tailed t-test was used for statistical analysis. P ≤ 0.05 was considered as statistically significant.
References
- "Yeast", 2012. http://dx.doi.org/10.1002/9783527659180
- D.E. Levin, "Cell Wall Integrity Signaling inSaccharomyces cerevisiae", Microbiology and Molecular Biology Reviews, vol. 69, pp. 262-291, 2005. http://dx.doi.org/10.1128/MMBR.69.2.262-291.2005
- G. Lesage, and H. Bussey, "Cell Wall Assembly in Saccharomyces cerevisiae", Microbiology and Molecular Biology Reviews, vol. 70, pp. 317-343, 2006. http://dx.doi.org/10.1128/MMBR.00038-05
- C. Cappellaro, V. Mrsa, and W. Tanner, "New potential cell wall glucanases of Saccharomyces cerevisiae and their involvement in mating.", Journal of bacteriology, 1998. http://www.ncbi.nlm.nih.gov/pubmed/9748433
- J. Zeitlinger, I. Simon, C.T. Harbison, N.M. Hannett, T.L. Volkert, G.R. Fink, and R.A. Young, "Program-Specific Distribution of a Transcription Factor Dependent on Partner Transcription Factor and MAPK Signaling", Cell, vol. 113, pp. 395-404, 2003. http://dx.doi.org/10.1016/S0092-8674(03)00301-5
- S. Bidlingmaier, E.L. Weiss, C. Seidel, D.G. Drubin, and M. Snyder, "The Cbk1p Pathway Is Important for Polarized Cell Growth and Cell Separation in Saccharomyces cerevisiae", Molecular and Cellular Biology, vol. 21, pp. 2449-2462, 2001. http://dx.doi.org/10.1128/MCB.21.7.2449-2462.2001
- M. Doolin, A.L. Johnson, L.H. Johnston, and G. Butler, "Overlapping and distinct roles of the duplicated yeast transcription factors Ace2p and Swi5p", Molecular Microbiology, vol. 40, pp. 422-432, 2001. http://dx.doi.org/10.1046/j.1365-2958.2001.02388.x
- R. Teparic, "Increased mortality of Saccharomyces cerevisiae cell wall protein mutants", Microbiology, vol. 150, pp. 3145-3150, 2004. http://dx.doi.org/10.1099/mic.0.27296-0
- A.F. Slater, C.S. Nobel, E. Maellaro, J. Bustamante, M. Kimland, and S. Orrenius, "Nitrone spin traps and a nitroxide antioxidant inhibit a common pathway of thymocyte apoptosis.", The Biochemical journal, 1995. http://www.ncbi.nlm.nih.gov/pubmed/7702573
- S. Jiang, J. Cai, D.C. Wallace, and D.P. Jones, "Cytochrome c-mediated Apoptosis in Cells Lacking Mitochondrial DNA", Journal of Biological Chemistry, vol. 274, pp. 29905-29911, 1999. http://dx.doi.org/10.1074/jbc.274.42.29905
- F. Madeo, E. Fröhlich, M. Ligr, M. Grey, S.J. Sigrist, D.H. Wolf, and K. Fröhlich, "Oxygen Stress: A Regulator of Apoptosis in Yeast", The Journal of Cell Biology, vol. 145, pp. 757-767, 1999. http://dx.doi.org/10.1083/jcb.145.4.757
- C. Fleury, B. Mignotte, and J. Vayssière, "Mitochondrial reactive oxygen species in cell death signaling", Biochimie, vol. 84, pp. 131-141, 2002. http://dx.doi.org/10.1016/S0300-9084(02)01369-X
- H. Eisler, K. Fröhlich, and E. Heidenreich, "Starvation for an essential amino acid induces apoptosis and oxidative stress in yeast", Experimental Cell Research, vol. 300, pp. 345-353, 2004. http://dx.doi.org/10.1016/j.yexcr.2004.07.025
- R. Del Carratore, C. Della Croce, M. Simili, E. Taccini, M. Scavuzzo, and S. Sbrana, "Cell cycle and morphological alterations as indicative of apoptosis promoted by UV irradiation in S. cerevisiae", Mutation Research/Genetic Toxicology and Environmental Mutagenesis, vol. 513, pp. 183-191, 2002. http://dx.doi.org/10.1016/S1383-5718(01)00310-2
- Q. Ren, H. Yang, M. Rosinski, M.N. Conrad, M.E. Dresser, V. Guacci, and Z. Zhang, "Mutation of the cohesin related gene PDS5 causes cell death with predominant apoptotic features in Saccharomyces cerevisiae during early meiosis", Mutation Research/Fundamental and Molecular Mechanisms of Mutagenesis, vol. 570, pp. 163-173, 2005. http://dx.doi.org/10.1016/j.mrfmmm.2004.11.014
- R.V. Pérez-Gallardo, L.S. Briones, A.L. Díaz-Pérez, S. Gutiérrez, J.S. Rodríguez-Zavala, and J. Campos-García, "Reactive oxygen species production induced by ethanol inSaccharomyces cerevisiaeincreases because of a dysfunctional mitochondrial iron-sulfur cluster assembly system", FEMS Yeast Research, vol. 13, pp. 804-819, 2013. http://dx.doi.org/10.1111/1567-1364.12090
- Q. Gao, Q. Ren, L. Liou, X. Bao, and Z. Zhang, "Mitochondrial DNA protects against salt stress‐induced cytochrome c‐mediated apoptosis in yeast", FEBS Letters, vol. 585, pp. 2507-2512, 2011. http://dx.doi.org/10.1016/j.febslet.2011.06.034
- D.K. Woo, T.L. Phang, J.D. Trawick, and R.O. Poyton, "Multiple pathways of mitochondrial-nuclear communication in yeast: Intergenomic signaling involves ABF1 and affects a different set of genes than retrograde regulation", Biochimica et Biophysica Acta (BBA) - Gene Regulatory Mechanisms, vol. 1789, pp. 135-145, 2009. http://dx.doi.org/10.1016/j.bbagrm.2008.09.008
- V. Mrsa, F. Klebl, and W. Tanner, "Purification and characterization of the Saccharomyces cerevisiae BGL2 gene product, a cell wall endo-beta-1,3-glucanase.", Journal of bacteriology, 1993. http://www.ncbi.nlm.nih.gov/pubmed/8458852
- J. Shimizu, K. Yoda, and M. Yamasaki, "The hypo-osmolarity-sensitive phenotype of the Saccharomyces cerevisiae hpo2 mutant is due to a mutation in PKC1, which regulates expression of beta-glucanase.", Molecular & general genetics : MGG, 1994. http://www.ncbi.nlm.nih.gov/pubmed/8152414
- L. Benov, L. Sztejnberg, and I. Fridovich, "Critical evaluation of the use of hydroethidine as a measure of superoxide anion radical", Free Radical Biology and Medicine, vol. 25, pp. 826-831, 1998. http://dx.doi.org/10.1016/S0891-5849(98)00163-4
- M. Weinberger, A. Mesquita, T. Caroll, L. Marks, H. Yang, Z. Zhang, P. Ludovico, and W.C. Burhans, "Growth signaling promotes chronological aging in budding yeast by inducing superoxide anions that inhibit quiescence.", Aging, 2010. http://www.ncbi.nlm.nih.gov/pubmed/21076178
- G. Farrugia, and R. Balzan, "Oxidative Stress and Programmed Cell Death in Yeast", Frontiers in Oncology, vol. 2, 2012. http://dx.doi.org/10.3389/fonc.2012.00064
- D. Carmona-Gutierrez, T. Eisenberg, S. Büttner, C. Meisinger, G. Kroemer, and F. Madeo, "Apoptosis in yeast: triggers, pathways, subroutines", Cell Death & Differentiation, vol. 17, pp. 763-773, 2010. http://dx.doi.org/10.1038/cdd.2009.219
- K. Fröhlich, and F. Madeo, "Apoptosis in yeast: a new model for aging research", Experimental Gerontology, vol. 37, pp. 27-31, 2001. http://dx.doi.org/10.1016/S0531-5565(01)00177-2
- A. Wach, A. Brachat, R. Pöhlmann, and P. Philippsen, "New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae.", Yeast (Chichester, England), 1994. http://www.ncbi.nlm.nih.gov/pubmed/7747518
- D.M. Gelperin, M.A. White, M.L. Wilkinson, Y. Kon, L.A. Kung, K.J. Wise, N. Lopez-Hoyo, L. Jiang, S. Piccirillo, H. Yu, M. Gerstein, M.E. Dumont, E.M. Phizicky, M. Snyder, and E.J. Grayhack, "Biochemical and genetic analysis of the yeast proteome with a movable ORF collection", Genes & Development, vol. 19, pp. 2816-2826, 2005. http://dx.doi.org/10.1101/gad.1362105
- H. Yang, Q. Ren, and Z. Zhang, "Chromosome or chromatin condensation leads to meiosis or apoptosis in stationary yeast (Saccharomyces cerevisiae) cells", FEMS Yeast Research, vol. 6, pp. 1254-1263, 2006. http://dx.doi.org/10.1111/j.1567-1364.2006.00123.x
- Q. Gao, Y. Liu, M. Wang, J. Zhang, Y. Gai, C. Zhu, and X. Guo, "Molecular cloning and characterization of an inducible RNA-dependent RNA polymerase gene, GhRdRP, from cotton (Gossypium hirsutum L.)", Molecular Biology Reports, vol. 36, pp. 47-56, 2007. http://dx.doi.org/10.1007/s11033-007-9150-y
SUPPLEMENTAL INFORMATION
![]() Download Supplemental Information
Download Supplemental Information
ACKNOWLEDGMENTS
This study was supported in part by an Institutional Development Award (IDeA) from the National Institute of General Medical Sciences of the National Institutes of Health under grant number P30 GM103398, by the National Natural Science Foundation of China (No. 31300070), and the China Postdoctoral Science Foundation (No. 2012M520850).
COPYRIGHT
© 2014

Salt stress causes cell wall damage in yeast cells lacking mitochondrial DNA by Qiuqiang Gao et al. is licensed under a Creative Commons Attribution 4.0 International License.