Viewpoint:
Microbial Cell, Vol. 3, No. 8, pp. 302 - 328; doi: 10.15698/mic2016.08.516
Similar environments but diverse fates: Responses of budding yeast to nutrient deprivation.
Division of Cell Biology and Biophysics, University of Missouri-Kansas City, 5007 Rockhill Rd, Kansas City MO 64110, USA.
Keywords: pseudohyphal growth, sporulation, meiosis, quiescence, Boolean logic, cell-cell signals, yeast communities.
Abbreviatons:
DPEB – differential patitioning provides environmental buffering,
FC – fermentable carbon source,
NFC – non-fermentable carbon source,
PCD – programmed cell death,
phg – pseudohyphal growth,
Q – quiescent,
ROS – reactive oxygen species,
SEDF – similar environment, different fate.
Received originally: 25/03/2016 Received in revised form: 07/07/2016
Accepted: 21/07/2016
Published: 01/08/2016
Correspondence:
Saul M. Honigberg, honigbergs@umkc.edu
Conflict of interest statement:
I declare no conflict of interest with the content of my article entitled “Similar environments but diverse fates: Responses of budding yeast to nutrient deprivation”.
Please cite this article as: Saul M. Honigberg (2016). Similar environments but diverse fates: Responses of budding yeast to nutrient deprivation. Microbial Cell 3(8): 302-328.
Abstract
Diploid budding yeast (Saccharomyces cerevisiae) can adopt one of several alternative differentiation fates in response to nutrient limitation, and each of these fates provides distinct biological functions. When different strain backgrounds are taken into account, these various fates occur in response to similar environmental cues, are regulated by the same signal transduction pathways, and share many of the same master regulators. I propose that the relationships between fate choice, environmental cues and signaling pathways are not Boolean, but involve graded levels of signals, pathway activation and master-regulator activity. In the absence of large differences between environmental cues, small differences in the concentration of cues may be reinforced by cell-to-cell signals. These signals are particularly essential for fate determination within communities, such as colonies and biofilms, where fate choice varies dramatically from one region of the community to another. The lack of Boolean relationships between cues, signaling pathways, master regulators and cell fates may allow yeast communities to respond appropriately to the wide range of environments they encounter in nature.
INTRODUCTION
A) Alternative cell fates: Biology ain’t always Boolean
Most tissues contain multipotent stem cells– i.e. cells able to differentiate into one or more cell types. The choice between fates depends largely on stimuli from the environment/niche of the cell. Often a given fate choice depends on multiple signals – some that promote and some that inhibit a particular fate. These observations suggest that Boolean logic may apply to cell-fate choice—i.e. a particular fate is adopted in response to the presence or absence of a particular combination (or combinations) of signals. For example, the lactose operon is active only when lactose is present and glucose is absent [1]. However, many fate choices do not fit easily into the framework of Boolean logic. Given the facile genetics and unmatched gene annotation in Saccharomyces cerevisiae [2], this yeast has served for many years as a model for the regulation of differentiation [3][4]. The present review focuses on fate choices in diploid cells of the budding yeast, S. cerevisiae (Baker’s yeast). I propose that many aspects of this choice are non-Boolean in nature.
–
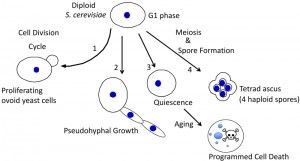
Diploid yeast can differentiate in multiple ways (Fig. 1). In particular, as nutrients become depleted, these cells differentiate in at least three distinct ways: 1) they can sporulate to form haploid spores (reviewed in [5][6]), 2) they can switch into “pseudohyphal growth” (phg) to grow as elongated chains of cells (reviewed in [7][8][9]), or 3) they can enter a stable non-proliferative state known as “quiescence” or “stationary phase” where they age and eventually undergo programmed cell death (reviewed in [10][11][12].
The current review focuses on the mechanisms by which S. cerevisiae chooses between these several “nutrient-deprivation” fates and the biological functions of each choice. Because in nature individual yeast cells typically proliferate, differentiate, age and die all within the context of multicellular communities such as colonies and biofilms, a particular focus of this review is how cell-fate decisions occur within these communities.
–
B) Central hypothesis: Similar environment – different fates
The central hypothesis presented in this review is that the choice of cell fate of S. cerevisiae is determined by relatively small differences in nutrient environment, which are then reinforced by cell-cell signals. I term this central hypothesis the “similar environment, different fate (SEDF)” hypothesis.
–
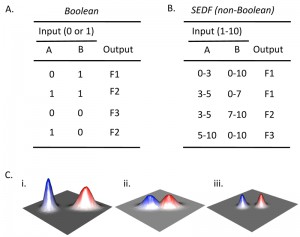
The SEDF hypothesis contrasts with a view in which each cell fate responds to discrete differences in environmental cues. Cell-fate decisions determined by discrete differences in cues can be expressed a Boolean relationship between these cues and a given cell fate. An example of a Boolean relationship between inputs and outputs is shown in Fig. 2A. Boolean logic requires that there are two states for each input (e.g. “1” and “0”) with respect to environmental cues. For example, if a response is linked to a threshold level (e.g. if a given fate requires the presence of a nutrient above a certain concentration), that would also be considered Boolean, since there are effectively only two states. An example of a non-Boolean relationship between input and output is shown in Fig. 2B. In this example the range of concentrations of a given cue that activate a particular cell fate depends on the concentration (not simply the presence or absence) of a second cue. Thus, the key feature of SEDF is that the relative level of multiple cues determines cells fate, not just their presence or absence.
Either a Boolean or non-Boolean model is consistent with the observation that each fate occurs most frequently in some environments than others. However, in a Boolean model, as mentioned above, the environments that promote one fate are clearly discrete from the environments that promote a different fate. A Boolean relationship is represented by a theoretical landscape (Fig. 2Ci). The discrete red and blue peaks in this figure represent two discrete differentiation responses; the two axes represent increasing intensity of two environmental cues (e.g. increasing concentration oxygen and nitrogen). In contrast, in a non-Boolean model the environments that promote each fate can overlap (Fig. 2Ci).
–
The landscape corresponding to the “red fate” in Fig. 2Ci shows an additional feature not allowed in Boolean models. Unlike the blue peak, in the red peak the two signals interact such that the response peak is not symmetrical relative to the axes. In other words, the optimal level for one cue is different, depending on the level of the second cue.
–
There are three main reasons to propose yeast cell fate follows an SEDF (non-Boolean) rather than a Boolean model, as discussed throughout this review. First, all 3 types of diploid differentiation occur in very similar environments, so fate choice is probably determined not by the presence or absence of one or more extracellular signals, but by the relative amount of these signals, i.e. fate choice cannot be represented by Boolean logic. Second, the transcription factors and signal transduction pathways that regulate yeast cell differentiation are not, either alone or in combination, specific for only one form of differentiation. Indeed, not only is the relationship between fate choice and environmental cues not Boolean, neither is the relationship between fate choice and the activity of most regulators. Third, yeast differentiation fates, despite mechanisms ensuring their stability, are remarkably flexible without dramatic changes in environment. For example, many communities of yeast are partitioned into populations undergoing different fates.
–
The main ramification of the SEDF hypothesis is to underline the importance of cell-cell signals in the context of yeast communities. As proposed below, the key mechanism that allows SEDF is cell-cell signaling that reinforces fate choice. Within these communities, the combination of environmental and cell-cell signals allows cell fate choice to be coordinated both temporally and spatially. In particular, I propose that relatively modest quantitative differences in nutrient environment (and signal pathways activity) are sufficient to efficiently specify a single cell fate because these small differences are reinforced by cell-to-cell signals. In the context of the theoretical landscapes described above, one can consider that these cell-cell signals allow discrete fate peaks even in a very similar environment—the effectively “sharpen” these peaks (Fig. 2Ciii). As a result, SEDF allows different regions of the community to adopt complementary fates within a relatively uniform environment. Furthermore, this type of coordination provides biological functions to the community that are not available to individual cells. As a result, these signals provide functions that may echo the origins of communication on this planet.
LESSONS FROM THE WILD
Before considering the evidence for SEDF, it is useful to regard possible differences between yeast found in nature (in which fate choices evolved) and yeast in the laboratory (in which fate choice can be studied). Recently, it has become clear that natural populations of S. cerevisiae exist throughout the world [13][14][15] and that these natural populations are different from domesticated yeast populations such as industrial or vineyard yeast as well as from clinical yeast isolates [16][17]. In particular, the ability to isolate, genotype, and in many cases sequence the genome of these natural isolates of S. cerevisiae has revealed at least two important features of the evolution of cell fate choice in wild yeast – the stability of the diploid state and the diversity of ecological niches in which these yeast can be found.
–
A) Natural cerevisiae populations are homozygous diploids
Based on genome analysis of many wild yeast isolates, it is clear that the ploidy of S. cerevisiae isolated from natural environments is diploid rather than haploid or polyploid [18][19][20]. By way of contrast, in the lab S. cerevisiae readily proliferates as haploids, diploids or polyploids. Indeed, polyploidy (particularly tetraploidy) is very common among industrial and food-processing S. cerevisiae [21][22], and polyploidy and even aneuploidy may occur relatively frequently in response to environmental stress (reviewed in [23]).
–
The fact that haploid yeast have not been isolated from the wild does not necessarily mean that sporulation is rare in natural environments. Indeed, yeast strains isolated from nature generally sporulate and mate efficiently in the laboratory under a range of nutrient conditions [24][25][26]. However, any spores isolated from nature might not be recognized as such, since they would probably germinate, mate and form diploids as soon as first cultured in the lab.
–
There are several reasons that haploids are likely short-lived intermediates both in nature and when first cultured in the lab. In the first place, after sporulation is complete, the four haploid products of sporulation, two of each mating type, are held tightly together in an ascal sac – the remnant of the cell wall of the parent cell. As a result, once nutrients are restored, haploids of opposite mating type efficiently undergo mating with other spores from the same ascus (intra-ascal mating) to restore the diploid state. Furthermore, any haploids that fail to mate with their sister spores would likely mate soon after beginning to proliferate, because most wild yeast are homothallic, meaning that a dividing cell produces a daughter cell of the opposite mating-type from the mother cell, allowing subsequent mating between mother and daughter. Finally, diploid cells proliferate more rapidly than haploid cells under most conditions [27], allowing diploids to out-compete haploids in the wild. The greater stability of the diploid state in S. cerevisiae contrasts with other yeast species such as S. pombe and Candida lusitaniae, which are both more stable in the haploid state. In these species, meiosis closely follows mating, rather than the reverse as in S. cerevisiae [28][29].
–
One implication of intra-ascal mating in wild yeast is that inbreeding between sister spores is much more frequent than outbreeding of unrelated yeast [30]. Also contributing to the low rates of outcrossing is the fact that yeast grows and sporulates primarily in clonal communities, i.e. starting from a single cell. Consistent with high levels of inbreeding in natural populations, analysis of molecular polymorphisms including whole-genome sequencing of natural isolates reveal them to be largely homozygous. Indeed, natural yeast have significantly less heterozygosity than clinical or vineyard isolates [31][32] perhaps because of less selection pressure from human associations on the natural yeast. Although inbreeding may be the rule for wild yeast, genome-wide sequencing indicates that outbreeding does occur [30] including between wild and domesticated yeast [13]. For example, outbreeding may occur in the gut of insect vectors such as wasps [33]. In fact, it has been suggested that the ratio of inbreeding to outbreeding can be regulated in wild yeast [34].
–
Haploids may be rare in the wild, but their ability to proliferate in the laboratory is extremely useful. Haploids can be easier to work with than diploids; for example, only a single allele must be deleted to eliminate the gene product. Furthermore haploids undergo many of the same fate choices as diploid cells. For example, haploids can switch from budding to filamentous growth, and haploids can enter quiescence and age. Although wild-type haploids are not normally able to initiate meiosis & sporulation, introduction of certain mutant alleles can bypass these constraints [35][36].
–
To return to the evolution of fate choice, since yeast found in the wild are almost always largely homozygous diploids, it is clear that the nexus for yeast differentiation is the diploid cell as nutrients become depleted. Below I describe how both yeast’s metabolism and its natural habitats provide the environmental cues that trigger differentiation.
–
B) Natural Saccharomyces habitats are both diverse and changeable
It is useful to consider the natural habitats/ecological niches of yeast in the wild with respect to the biology of fate choice. In the lab, yeast differentiate when nutrients becoming limited, not only because nutrient depletion slows or halts proliferation, but also because it directly promotes differentiation. As discussed in this section, it is particularly the changes in metabolism and hence nutrient environment accompanying late stages of growth that promote differentiation.
–
The crux of S. cerevisiae metabolism is the “Crabtree effect” [37]. The Crabtree effect describes the observation that when glucose is available, yeast will metabolize (ferment) this sugar completely to ethanol, acetate and other non-fermentable carbon sources (NFCs), and these non-fermentable products are only themselves efficiently metabolized further (via respiration) once glucose is completely exhausted. This switch from fermentation to respiration, termed the diauxic shift, occurs even when oxygen remains plentiful throughout. Not surprisingly, while glucose is plentiful, multiple signal transduction pathways respond to this glucose to repress respiratory enzymes and metabolism of alternative carbon sources (reviewed in [38]). Thus, S. cerevisiae is primarily adapted to proliferate on glucose and convert it to non-fermentable carbon.
–
Many yeast species do not display the Crabtree effect, and it has been suggested that this effect evolved in S. cerevisiae and other “Crabtree yeast” as a mechanism to out-compete surrounding microbial species because the ethanol produced by fermentation and secreted by the yeast inhibits the growth of these other species [39]. The Crabtree effect is also central to the economic power of S. cerevisiae because it allows the production of high levels of ethanol during fermentation and high levels of CO2 during respiration.
–
Wild yeast have been observed to proliferate in nature primarily on plant matter rich in sugars, such as tree exudates or rotting fruit [14][26]. In contrast, recent isolation of S. cerevisiae from sites throughout the world such as soil demonstrates that this species is capable of inhabiting a wide range of ecological niches that are not rich in glucose [15][40]. It seems likely that in these latter niches, yeast is not proliferating and may exist primarily as spores.
–
Nutrient environment changes rapidly both during late stages of growth and during differentiation itself. During late stages of growth, fermentable carbon sources, nitrogen and other nutrients become depleted whereas NFCs can remain relatively plentiful (Fig. 3). Indeed, both the intracellular and extracellular metabolome studies reveal dramatic changes in the concentrations of intermediary metabolites and amino acids during the diauxic shift [41][42]. These changes continue as differentiation progresses. For example, analysis of sporulation in transcriptome [38][43], proteome [44][45], and metabolome [46][47] studies indicate that the expression and activity of metabolic enzymes continues to fluctuate as sporulation progresses.
In summary, the biology and natural ecology of yeast indicate that yeast is distributed widely, but that it may primarily proliferate on fermentable carbon sources (FCs). The Crabtree effect ensures that as cells begin to exhaust nutrients and slow or cease growth, NFCs remain relatively plentiful. As discussed in the Section “Shared environmental cues & distinct fates”, this nutrient environment is optimal for differentiation regardless of the particular fate (consistent with SEDF). Thus, both the metabolism and the ecology of yeast suggest that yeast evolved such that late stages of growth provide the environmental cues necessary for differentiation.
–
C) A caveat regarding comparing laboratory and natural strains
Regulation of cell differentiation by nutrient environment has been studied in many different laboratory strains of yeast. It is now evident that these strain backgrounds can vary significantly with respect to the relationship between environmental cues and differentiation fates. These results raise the question of how well fate choice in lab strains reflects the fate choices that evolved in the wild. Indeed, strain variants with altered differentiation responses may have been selected in early laboratory strains, descendants of which are now used in most modern laboratories [48]. The effect of strain backgrounds on differentiation responses is a critical consideration when synthesizing data from different studies done in different strain backgrounds.
–
One example of phenotypic variation between common laboratory strain backgrounds is sporulation efficiency. Moreover, this variation extends to different isolates of industrial, clinical and wild yeast. Among some of these strains, variation in sporulation efficiency has been traced (e.g. by QTL analysis) to allele differences at a relatively few loci, e.g. the transcription factor Rme1 [32][49][50]. Furthermore, sporulation efficiency under a single condition likely underestimates the variation in sporulation capacity between strains. As one example, several common lab strain backgrounds (e.g. W303 and SK1) sporulate very efficiently under optimal conditions but sporulate much less efficiently than natural isolates on low concentrations of glucose [25].
–
Phg efficiency also varies considerably between strain backgrounds – both among laboratory strain backgrounds and among natural isolates of yeast [51]. In the strains that have been compared, this variation is again largely attributable to one or a few loci; for example, the transcription factor Flo8, a master regulator of phg [52][53][54]. In fact, several common laboratory strain backgrounds of yeast that lack a functional allele of Flo8 are completely unable to undergo phg, but can partially recover this ability when Flo8 is restored [55]. As with sporulation, variation in phg extends beyond efficiency under optimal conditions. For example, in some laboratory strain backgrounds (e.g. Ʃ1278b) phg occurs most efficiently in medium containing glucose, whereas in other strain backgrounds (e.g. SK1), phg occurs most efficiently in NFCs [56][57][58][59]. Finally, as with phg and sporulation, the rate of ageing in quiescent cells varies significantly between different laboratory strains [60].
–
In addition to allele variation for master regulatory genes such as RME1 and FLO8, the responsiveness of signal transduction pathways to environmental cues varies significantly between laboratory strains (reviewed in [61]). Many experiments connecting signaling and differentiation pathways have been done in only a single strain background, so it is wise to be circumspect in synthesizing results based in different strain backgrounds. An equally important point is that it is unlikely that any single laboratory strain background represents the “real” evolved response; indeed, the variation between laboratory strains is mirrored by variation between natural isolates.
–
Obviously, experiments with lab yeast strains have driven and will continue to drive most of what we understand about fate choice, as they do for most yeast biology. However, in considering the implications of these experiments to fate choice, it is useful to remember possible differences between these lab strains and the natural yeast strains in which fate choice evolved.
VARYING FATES, VARYING FUNCTIONS
From a functional viewpoint, a key aspect of the choice between cell fates is that each fate has a different biological role. In this section, these roles are discussed, especially in the context of the nutrient limitation that triggers diploid yeast differentiation.
–
A) Fate #1: sporulation – sex, food and energy
The primary function of sporulation is to produce cells (haploid spores) that are more resistant to environmental stresses than the vegetative cells from which they derive. For example, spores resist antimicrobials, high temperatures and prolonged starvation to a much greater extent than vegetative cells [62][63][64]. This resistance derives in large part from the thick walls encasing each spore [65][66]. Another aspect of spore resistance is that spores survive the insect gut much better than vegetative or quiescent cells [62]. Fruit flies and other insects are thought to be a major vector by which yeast spread from one ecological niche to another and outbreed to less related strains [67][68]. Indeed, it has been proposed that the environment of the insect gut is the primary site at which the spore wall protects viability [5].
–
Another presumed function of sporulation is to increase genetic diversity in a yeast population as a result of meiotic recombination coupled with independent assortment of chromosomes. Indeed, meiotic recombination in yeast may also increase genetic variation as a result of increased mutation rates near meiotic recombination sites [69]. However, as mentioned earlier, most wild yeast communities are clonal populations of largely homozygous strains. Yeast meiosis could be important in generating diversity after relatively rare out-breeding or mutation events by stimulating loss-of-heterozygosity at new alleles. In any case, yeast meiosis likely provides a relatively minor selective advantage relative to the increased environmental resistance of spores.
–
Sporulation as a response to limiting nutrients is particularly interesting in the context of the large energy requirement for the sporulation program. For example, energy is required to express hundreds of gene products required for meiosis and spore wall formation including many, such as spore wall proteins, that are produced to very high levels [44][45][70][71][72]. In addition, many of the cellular processes required in sporulation have additional energy requirements—e.g. DNA replication and chromosome segregation. This abundant expenditure of energy in response to nutrient limitation has been termed the sporulation “energy paradox” [73][74].
–
It is likely that this apparent paradox is resolved through a combination of several mechanisms. In the first place, storage carbohydrates, for example glycogen, accumulate during late stages of growth and are subsequently utilized for energy during sporulation (reviewed in [75]). Indeed, mutants defective in accumulating these storage carbohydrates fail to sporulate [76][77]. In the second place, deprivation for nitrogen (or other essential nutrients) when NFC is still abundant allows abundant energy production/respiration in the absence of cell division. Thus, yeast build environment-resistant spore walls by using both internal and external energy sources generated during growth.
–
B) Fate #2: Quiescence – dormancy, ageing & death
1) Not dead, just quiet. Quiescence is a differentiated state in which yeast cease growth (reviewed in [12]) and undergo genome-wide changes in transcriptional expression and chromosome topography [78][79][80], cytoskeletal organization [81] and cytosolic fluidity [82][83]. Thus, like sporulation, quiescence is a response to nutrient deprivation that is likely to require an energy investment. One of the major functions of quiescence is the same as that of sporulation – resistance to environmental stresses. For example, activation of the Mpk1 cell wall integrity pathway in quiescent (Q) cells induces the induction of cell-wall repair genes [84][85], and activation of Rim15 kinase in these same cells induces stress-resistance genes [86][87][88]. Quiescence in yeast has been studied primarily in haploids but is equally available to diploid cells.
–
Interestingly, Q diploids are more resistant to environmental stress than growing cells but less resistant than spores to environmental stress [5]. Q cells do not remain viable indefinitely. As time passes and Q cells age, their viability diminishes. Thus, quiescence, aging, and eventual death can be considered progressive stages in a single differentiation pathway.
–
2) The universal fate choice— getting older vs. the alternative. As stated above, cell death occurs naturally as cells age. For example, in suspended yeast cultures most cells have reached the end of their lifespan approximately 1-3 weeks after they have ceased growth [63][89][90]. Lifespan is ended through a programmed cell death (PCD) (reviewed in [91][92][93]).
–
PCD in yeast displays many of the same cellular landmarks as apoptosis in higher organisms, such as DNA fragmentation, cell surface changes, and involvement of mitochondria and reactive oxygen species (ROS) [94][95]. For this reason, yeast PCD is often referred to as “yeast apoptosis”. However, yeast PCD does not utilize all of the same regulators as mammalian apoptosis nor the extensive family of caspases typical of metazoan apoptosis [96][97]. In this review, to avoid semantic distinctions, I refer to yeast apoptosis as PCD.
–
Yeast lifespan is limited not only by the period of time that elapses after growth ceases (chronological ageing), but also by the number of times a mother cell can divide before it dies (replicative ageing) (reviewed in [98]). These two types of ageing are regulated by many of the same pathways (reviewed in [99][100]), and they are also linked in the sense that chronologically aged yeast have shorter replicative lifespans than chronologically young cells [101]. Nevertheless, the two types of age are not interchangeable; for example, the Sir2 histone deacetylase inhibits replicative ageing, but Sir2 actually stimulates chronological ageing in some strain backgrounds and conditions (reviewed in [102]).
In addition to ageing, yeast PCD is also triggered by a wide variety of other environmental stresses [103][104][105][106]. A common feature between ageing and most other triggers for PCD in yeast is the accumulation of oxidative and other cellular damage. According to one view of ageing, accumulation of damage over time eventually triggers PCD (reviewed in [107]).
–
What is the relationship of nutrient environment to ageing and subsequent cell death? A common feature of replicative lifespan control from yeast to metazoans is that lifespan is increased when nutrients are limited, i.e. calorie restriction [108][109]. Although the role of calorie restriction in lifespan extension is still a matter of debate, one idea is that when metabolism is limited, ROS and hence oxidative damage are also limited, and as a result lifespan is extended [110][111][112][113]. A corollary to this hypothesis is that inducing repair of oxidative stress also extends lifespan [114][115].
–
Regardless of the of PCD trigger, its function in yeast and other single-celled organisms has been the subject of debate. One idea is that because yeast growth is largely clonal, programmed death in one cell could benefit other cells with the same genotype, thus providing a selective advantage for the genotype. For example, suspended cultures accumulate ROS after prolonged incubation, triggering PCD in most of the culture and presumably releasing enough nutrients from the dying cells to allow a subpopulation of still viable cells to continue growth and acquire adaptive mutations [116]. More generally, several investigators have proposed mechanisms by which programmed cell death benefits the overall (or average) survival of a clonal community in S. cerevisiae [92][117] and other microorganisms [118][119][120]. The specific case of cell fate choice (including PCD) within communities is discussed further in the section “Shared communities – coordinated fates”.
–
In summary, quiescence, ageing and PCD can be considered a single progressive pathway with different functions at earlier stages (e.g. resistance to environmental stress) than at later stages (e.g. possible re-distribution of nutrients).
–
C) Fate # 3: Phg- the life of the forager
1) Overview: looking for a better neighborhood. Phg, like both quiescence and sporulation, is a response to diminished nutrients. However, phg is unique among yeast diploid differentiation fates in that it is also a means of cell proliferation.
–
One likely function of phg is foraging. Specifically, phg allows yeast communities such as colonies to expand and access distant nutrients more efficiently than is possible when yeast divide through its standard (ovoid) budding patterning. Indeed, in many strain backgrounds phg occurs only in communities, not in suspended cultures [121]. However, in other strain backgrounds pseudohyphae will form under certain conditions even in suspended cultures [122][123].
–
At least two types of foraging are associated with pseudohyphae: i) extension of chains of elongated yeast cells along the surface of the underlying substrate, and ii) invasion of these chains into the underlying substrate.
–
2) Exploring the surface. The first type of foraging is closely related to the structure of pseudohyphae as chains of elongated cells. These chains radiate out from the perimeter of a colony or biofilm along the surface of the agar or other hard surface like plastic on which these communities grow. Limiting phg to the fringe of the community may be the most efficient mechanism to access distant sources of food.
–
3) Exploring below the surface. As yeast colonies mature, they sometimes grow into (or “invade”) the underlying agar medium. Although invasive growth is often studied in haploids, diploids colonies are equally capable of invading agar. By the same reasoning as above, invasive growth potentially allows access to distal nutrients. In addition, invasive growth may provide benefits by anchoring a yeast community to its underlying substrate, and invasive growth of yeast into fruit and other natural substrates have been observed [124]. By analogy, the much longer hyphal and pseudohyphal filaments formed by the pathogenic yeast Candida albicans are necessary for these yeast to invade host tissue and, hence, for pathogenicity (reviewed in [125]).
–
In theory, the elongated-chain geometry of pseudohyphae could provide the force necessary for invasion, but in fact the connection between pseudohyphae and invasive growth is not straightforward. Most laboratory strains that can form pseudohyphae can also invade agar [57][59][126]. However, many genes have been implicated in one program but not the other [127][128]. Furthermore, in some wild and laboratory yeast grown on agar plates, the region where colonies invade the agar is not associated with extensive phg [25].
–
4) Phg and phg-spectrum phenotypes. Invasiveness is only one of a spectrum of wild-type phenotypes that require many of the same genes as phg but which do not always directly require phg. In particular, these “phg-spectrum” phenotypes all depend on expression of flocculins, which are a class of lectin-type proteins involved in both cell adhesion and cell signaling [129]. The diversity of flocculin-dependent phenotypes reflects the variety of communities that can form in this species. For example, the FLO11 flocculin is required to form “structured colonies”, which have with a distinctive lacey appearance [130][131], “mats”, which are large thin colonies formed on low agar (high moisture) plates [132], “flors”, which are thin colonies of yeast that form on the top of liquid cultures [133], “flocs”, which are large clumps of cells that form suspended in cultures [58][134], and “minicolonies” which are biofilm like structures that adhere to plastic surfaces submerged in medium [56][133]. Both lab and natural yeast isolates vary considerably in their ability to undergo these phg-spectrum phenotypes [20][131]
–
D) Summary – distinct functions for discrete fates
As discussed in the Section “Lessons from the wild”, all three differentiation fates are a response to a limited-nutrient environment, but as we saw in the Section “Varying fates, varying functions”, each fate has distinct functions and costs from the other two cell fates. For example, both quiescence and sporulation result in cells that are more resistant to the environment, but the higher resistance of spores relative to quiescence comes at the expense of a sizeable energy investment. Phg, a response to less severe nutrient depletion than the other two fates, functions more for foraging than for resistance to stress. Because fate choices are functionally quite distinct, it is striking, as discussed below, that they all respond to many of the same environmental cues, signaling pathways, and master regulators.
OVERLAPPING REGULATORS BUT DISTINCT FATES
A) Introduction to the master regulators of differentiation
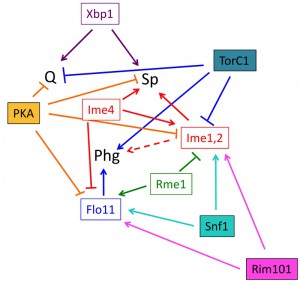
At one level, differentiation fate is determined by expression of master regulator(s). Master regulators are gene products that are required to initiate differentiation; indeed by one definition, a true master regulator is sufficient to trigger differentiation when expressed ectopically under conditions where that fate is normally suppressed. In a Boolean logic system, each fate would be defined by the presence or absence of one or more master regulators. Indeed, each differentiation pathway in yeast requires one or more master regulators and subsequent expression changes in hundreds of genes. However, as described below several of these master regulators activate more than one differentiation pathway, repress more than one pathway, or activate one pathway while repressing another (Fig. 4).
Phg is regulated by a set of transcription factors including Flo8, Ste12, Tec1 and Nrg1, all of which activate transcription of the flocculin gene FLO11 (reviewed in [135]). Sporulation is initiated by expression of the Ime4 RNA methylase, the Ime1 transcription factor and the Ime2 protein kinase (reviewed in [5]). Quiescence is activated by expression of the Xbp1 transcription factor [78][79].
–
B) Activators of meiosis (IME1, IME2) also activate phg
Surprisingly, two key master regulators of sporulation, the Ime1 transcription factor and Ime2 kinase, are also required for phg in some laboratory strain backgrounds. In particular, these genes are required for phg in the SK1 but not the Ʃ1278b background [57]. This result is consistent with an observation described above—in SK1, phg (like IME1 and IME2 expression) is induced by NFC, but in Ʃ1278b phg is induced by glucose, which strongly inhibits IME1 and IME2.
–
The roles of Ime2 in activating both phg and sporulation in SK1 is broadened further when Ime2 homologs in other fungal species are considered. For example, Ime2p homologs in Aspergillus nidulans and Neurospora crassa activate the development of sexual structures, and Ime2 homologs in Ustilago maydis and Cryptococcus neoformans activate mating and filamentous growth (reviewed in [136]). Thus, the idea that a master regulator can control more than a single differentiation program extends across species.
–
C) Rme1 and Ime4 activity and the sporulation/phg choice
Rme1 is a transcriptional activator that inhibits sporulation and stimulates phg. Rme1 prevents sporulation in haploid cells, a critical function since haploid meiosis is inevitably lethal. Rme1, which is expressed at high levels in haploids exposed to sporulation conditions, prevents meiosis by activating transcription of IRE1 a non-coding gene approximately 1 kb upstream of IME1. Transcription of IRE1 through the IME1 promoter prevents IME1 transcription [137].
–
At the same time that Rme1 blocks IME1 transcription in haploids, it also activates FLO11 transcription to promote filamentous growth [138]. Rme1 is also expressed in diploids (though at lower levels), and hence may balance sporulation and pseudohyphal fates in diploids. For example, wild yeast isolates with relatively high Rme1 expression tend to have low sporulation and high phg, whereas strains with relatively low Rme1 expression have the reverse tendency [32].
–
Not surprisingly, both IME1 and FLO11 are regulated by a number of other transcription factors in addition to Rme1. Indeed, IME1 and FLO11 have among the largest upstream intergenic distance of any yeast gene – consistent with these genes having especially complex promoters [139]. Multiple nutrient signals are integrated in regulating FLO11 not only through the transcription factors that bind its complex promoter but also through regulating activity of the Msb2/MAPK pathway that activates some of these transcription factors [135].
–
The expression pattern of IME4 is opposite to that of RME1 — IME4 is expressed to higher levels in diploids than in haploids [140]. Like IME1, IME4 is regulated by an overlapping long noncoding transcript [141][142]. Furthermore, Ime4 also acts opposite to Rme1 in balancing sporulation with phg. For example, Ime4 inhibits FLO11 transcript accumulation while promoting Ime1 transcript accumulation [141][143]. Ime4 is an RNA N-6 adenosine methyltransferase that likely acts on many hundreds of RNAs [144]. Interestingly, N-6 adenosine methylation also regulates expression of large gene sets in plants and animals during differentiation and development (reviewed in [145]). Other proteins that (like Ime4) promote sporulation and inhibit phg include Bir1, which is homologous to IAP (inducer of apoptosis protein) [146], and Spo21, a 67 aa protein that localizes to the prospore [147].
–
D) Quiescence and sporulation: shared regulator & shared properties
Xbp1 represses transcription of many genes by targeting the Rpd3 histone deacetylase to these genes [79]. Interestingly, Xbp1 is not only required for quiescence but also for sporulation, at least in part because Xbp1 represses transcription of G1 cyclins [148][149]. These cyclins not only trigger the G1 to S transition, they also repress IME1 transcription and inhibit Ime1p nuclear localization [150][151]. It is not known whether Xbp1 is required for all forms of quiescence.
–
Most meiotic genes are not induced in quiescent cells, but at least some of the same metabolic enzymes and stress resistant enzymes are induced in both programs [46][152]. For example, trehalose synthesis is required for both initiation of meiosis and maintenance of quiescent cell viability [76][153]. Similarly, many proteins required for heat shock resistance are induced during both quiescence/ageing and sporulation [44][154][155]. It is possible that cells first enter quiescence and only then may sometimes also initiate sporulation. Alternatively, quiescence and sporulation may be mutually exclusive fates that share certain common regulators and target genes.
–
E) Summary
Master regulators of diploid cell fate do not display Boolean-logic relationships to fate choice, i.e. there is not a particular combination(s) of known master regulators that specify each fate (Fig. 4). Instead, the relationship between master regulators and cell fate is complex. Indeed, some regulators, such as Flo11 regulate only a single fate, whereas other regulators (Xbp1, Ime1, and Ime2) activate more than one fate. Finally, a third class of regulators (Rme1 and Ime4), activate one fate while repressing another. How these master regulators together determine fate choice is unknown, but evidence so far indicates that their relationship to fate choice is non-Boolean.
SHARED ENVIRONMENTAL CUES & DISTINCT FATES
A) Introduction: a common environment stimulates each fate
Given that all three diploid differentiation programs occur as nutrients become limiting, how does a cell choose between programs? It is possible that a small difference in the concentration of one or more nutrients under these conditions would result in passing a concentration threshold required for activation of a single fate choice. This would be an example of a cue/fate relationship that could be represented a Boolean logic since there are only two states relative to the threshold. This section addresses the question of whether the evidence allows for a Boolean relationship between cues and fate.
–
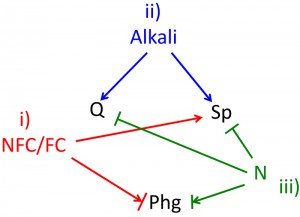
A summary of the relationships between environmental cues and differentiation pathways is shown in Fig. 3. Sporulation occurs under the specific condition of active respiration (NFCs being much higher than FCs), high pH, and depleted nitrogen and/or other nutrients (reviewed in [156][157]). Quiescence, like sporulation, is triggered by the absence of at least one essential growth nutrient and alkaline pH, but (unlike sporulation) quiescence does not have a requirement for respiration (reviewed in [10][158]. Finally, pseudohyphal differentiation occurs at intermediate-to-low nitrogen concentrations (reviewed in [135]), can also be induced by other cues such as fusel alcohols, and also responds to other cues in some strain backgrounds (reviewed in [159]).
–
B) Carbon source
Yeast can metabolize many different carbon sources, but from the point of view of cell differentiation, there are two main types of carbon source. The first type is the FCs, in particular glucose, which is fermented through glycolysis to produce the second class – the NFCs. During late stages of growth, NFCs, particularly ethanol and acetate, are metabolized to carbon dioxide, and the balance between glucose and NFC during late stages of growth is a critical determinant of cell fate (Fig. 3).
–
1) Carbon source, quiescence and sporulation. Glucose even in low concentrations effectively inhibits the initiation of meiosis in most laboratory strains. Presumably, sporulation has evolved such that it only initiates after the favored carbon source for growth, glucose, is fully metabolized to NFCs. For sporulation to initiate, a second cue besides the absence of glucose is the presence NFCs. Indeed, sporulation requires not only the energy provided by respiration but also the specific presence of a NFC [160], and the continued presence of NFCs is required even at late stages of sporulation [161]. Thus, as the ratio of NFCs to glucose (or other FCs) increases at late stages of growth, the environment becomes increasingly optimized for sporulation (Fig. 3).
–
Quiescence is classically defined as occurring after the complete depletion of both NFCs and FCs [12]. However, a core set of genes is induced during quiescence regardless of which nutrient (C, N or PO4) is limiting [162][163]. Although quiescence can be induced in a range of environments, extracellular environment strongly influences the properties of Q cells. For example, metabolic and other biological properties of Q cells vary depending on which nutrient is limiting (reviewed in [10][164]). These different properties suggest the existence of multiple types of Q cells depending on the presence or absence of particular environmental cues. For example, ethanol and acetic acid accelerate chronological ageing in Q cells [60][99][165][166].
–
2) Carbon & phg. A change in the ratio between NFCs and glucose likely regulates phg in all strain backgrounds, but the optimal NFC/glucose ratio may vary between backgrounds. For example, phg occurs efficiently in the Ʃ1278b background when grown on glucose medium, but is much more efficient in the SK1 background when grown on acetate medium [56][57]. Several results may help to explain this difference. In the first place, even in Ʃ1278b, high concentrations of glucose (2%) inhibit haploid filamentous and invasive growth [167][168]. In the second place, assays for phg in glucose medium generally involve cells dividing for many generations before N limitation triggers phg (termed the dimorphic switch) [169]. Thus, at least some glucose has been converted to NFCs by the time phg initiates. In this respect, increased NFC/glucose ratio during late stages of growth may stimulate phg as well as sporulation; though the optimal ratio may be lower in Ʃ1278b than in SK1.
–
C) Nitrogen supply
1) Nitrogen availability, growth and differentiation. It is tempting to neatly classify fate choice in yeast as driven by the presence or absence of two or three nutrients; yet, in nature yeast must make cell fate decisions across a range of different environments. The relative nitrogen availability from different sources illustrates this point. Yeast is capable of assimilating nitrogen from many different sources; however, the efficiency of utilizing nitrogen varies considerably depending on the source [170]. For example, ammonium and glutamine are very good nitrogen sources for growth, whereas urea and tryptophan are poor sources. As discussed below, nitrogen “quality”, besides affecting growth rate, also affects fate choice.
–
2) Nitrogen and fate choice. In laboratory cultures, nitrogen is often the nutrient that becomes limiting during late stages of growth. In addition to directly regulating differentiation (see Section “Same signal paths – different fates”), nitrogen limitation prevents sufficient G1 cyclin from accumulating to activate the G1-to-S transition (START). The START transition blocks both sporulation and quiescence, both of which can only initiate from G1. Furthermore, cyclin expression inhibits both transcription of IME1 [150][151][171] and nuclear localization of Ime1 [172]. Similarly, cyclin expression also blocks the establishment and maintenance of quiescence [79]. Finally, once cells do enter quiescence, higher quality N sources accelerate ageing relative to lower quality sources [173].
–
An intermediate level of nitrogen is required both for phg and many phg-spectrum phenotypes [59]. For example, FLO11 is induced when low concentrations of nitrogen are present in the medium but repressed both at high nitrogen concentrations and when nitrogen is completely absent [174]. Similarly, both FLO11 induction and phg require intermediate G1 cyclin levels [128]. Thus, phg requires both detecting and utilizing intermediate N concentrations [175][176][177].
–
In summary, nitrogen level/availability regulates cell fate, not just its presence or absence or its concentration relative to a single threshold. Thus, the role of nitrogen in regulating differentiation cannot be represented by a Boolean “1” vs. “0” relationship between this cue and cell fate.
–
D) The simplest of signals: pH
1) Extracellular pH fluctuates dramatically during growth. Changes in extracellular pH coincide with nutrient depletion and contribute to fate choice. Proliferation is much less sensitive to external pH than is differentiation – yeast grows efficiently through a broad pH range (pH 3-8) – in part because acid pumps provide a relatively uniform intracellular pH regardless of external pH [178]. However, extracellular pH does change dramatically during both proliferation and differentiation. During fermentative growth, the secretion of organic acids such as acetate and pyruvate decreases external pH ≤ 4.0 [166]. In contrast, in media containing plentiful nitrogen and other nutrients, subsequent respiration of these organic acids during the diauxic shift converts these acids to C02, which is either released as a gas or solubilized as bicarbonate. As a result, during the diauxic shift, extracellular pH increases to ≥ 8.0. In this respect, extracellular pH reflects the ratio of FCs to NFCs [179]. Note that if the ratio of glucose to other essential nutrients is high enough in the chosen growth medium, then glucose will never be exhausted, and cells will not undergo either the diauxic shift or the second pH transition.
–
2) Extracellular pH has complex effects on cell differentiation. Increased extracellular pH during late stages of growth stimulates quiescence [180]. Likewise, when Q cells are exposed to low pH, their viability (chronological lifespan) is strongly diminished [166][181]. Increased extracellular pH also activates sporulation, and this pH continues to rise as sporulation progresses [182][183]. Finally, pH has varying effects on phg-spectrum of phenotypes. For example, flocculation is stimulated by acidic pH [58][184][185], whereas, invasive growth is stimulated by alkaline pH [186].
–
E) Is it really just all about the food?
In natural environments, yeast must adapt to temperature fluctuations over the course of the day and the course of the year. In general, diploid differentiation is more sensitive to temperature than is proliferation. For example, most laboratory strains are unable to sporulate at moderately high temperatures (> 34°C) even though growth is still efficient at temperatures exceeding 37°C [187]. Similarly, many clinical yeast isolates are able to grow as ovoid cells at much higher temperatures than as pseudohyphae [188][189]. Finally, as cells enter quiescence, resistance to heat and other stress initially increases and then declines as cells age [190][191].
–
Another aspect of cellular environment is cell-cell contacts. The role of cell-cell contacts in cell-fate decisions is most clear for phg, where cell adhesion molecules like the flocculin, Flo11, and the mucin, Msb2, are required for many phg-spectrum phenotypes (reviewed in [192][193]). Sporulation in minicolonies might also be regulated by cell-surface contacts, since only the pseudohyphae projecting from minicolonies, not cells in the core, are capable of sporulating [56].
–
F) Summary
Given that all three types of diploid differentiation occur in the nutrient-depleted environment characteristic of late stages of growth, it is not surprising that the environmental cues that regulate them are similar, but there are two points worth emphasizing. First, although each pathway occurs independently, there is no single combination of the presence or absence of cues that unambiguously specifies a single fate (Fig. 3). Second, the level or relative concentration of an environmental cue (e.g. ratio of NFCs to FCs or low, intermediate or high nitrogen) often correlates better with fate choice than simply the presence of that cue above some single threshold concentration. Both findings support the SEDF hypothesis and argue against a Boolean relationship between cues and fates.
SAME SIGNAL PATHS – DIFFERENT FATES
A) Overview
It is striking that all three diploid differentiation pathways not only occur in similar nutrient environments but are also regulated by the same four major nutrient signal transduction pathways (PKA, TorC1, Snf1 and Rim101). As with master regulators and environmental cues, in the case of signaling pathways, one can imagine pathways in only one of two states, ON or OFF, relative to pathway targets including differentiation programs. Alternatively, there could be multiple or even continuous (graded) pathway activity levels. As one example, variable numbers of receptors could result in nearly continuous activity levels. The mechanisms and components of signal transduction have been discussed in several excellent recent reviews [3][61][194][195], so this section of the viewpoint focuses only on the role of these pathways in regulating fate choice and the possibility of a Boolean relationship between signal pathways and fate choice (Fig. 4).
–
B) Complex relationship between signal paths and nutrient signals
Study of most yeast nutrient signaling pathways initiated with a focus on the relationship between a single nutrient and a single pathway. For example, the TorC1 pathway is primarily activated by nitrogen, the Ras/PKA pathway is primarily activated by glucose and the Snf1 pathway is primarily repressed by glucose. However, it is now apparent that most nutrient-responsive pathways relay information about more than one type of nutrient. For example, multiple receptors responding to different classes of ligands can converge to regulate the same signaling pathway.
–
The Snf1 pathway exemplifies the ability of a single pathway to respond to diverse cues. Although this pathway was identified and characterized as active when glucose is absent, and is often referred to as the “glucose repression pathway”, it also responds to other types of cellular stress (reviewed in [196]). For example, in the absence of glucose, this pathway responds instead to nitrogen levels [197]. Similarly, the TorC1 pathway is not only activated by abundant nitrogen, but also by glucose, by osmotic stress, and by other types of cellular stress [198]. Finally, the PKA pathway, which is activated in high glucose through the Ras branch of the pathway, is also regulated through other branches that respond to carbon source, ammonium, amino acids and phosphate (reviewed in [199]). In fact, even the Ras branch of the PKA pathway is not regulated solely by glucose but is sensitive to acetate levels [200] and may respond more generally to cytosolic pH [201][202].
–
C) PKA inhibits all three forms of differentiation
PKA is active during rapid growth in FCs and has reduced activity in Q and sporulating cells. In yeast (and metazoans) PKA activates both the expression and activity of metabolic enzymes required for rapid growth. In addition, PKA represses genes required in non-proliferating, slow growing, and respiring cells (e.g. glycogen storage). Thus, PKA activity is low during all three types of cell differentiation.
PKA inhibits differentiation through both general mechanisms, which repress all three differentiation programs, and specific mechanisms, which repress only one (or two) of the three differentiation choices. One general mechanism for inhibition occurs when PKA phosphorylates Whi3 protein [203]. Once phosphorylated, Whi3 releases CLN3 mRNA, allowing progression of the cell cycle from G1 to S (START) and corresponding inhibition of quiescence and sporulation [203][204][205].
–
Another general mechanism by which PKA inhibits all three forms of differentiation is by phosphorylating and inactivating Rim15 kinase [206]. Rim15 is required for the metabolic changes that accompany quiescence [207] and also activates the endosulfine, Igo1, which helps maintain mRNA populations during quiescence [208][209][210]. Rim15 also activates the Msn2/Msn4 transcription factors, which directly activate stress response genes necessary for quiescence [86][211], and Rim15 is also required for the lifespan extension caused by calorie restriction [115][212]. In addition to its role in quiescence, Rim15 is required for both the expression and activity of Ime1 [213][214][215]. Thus, Rim15 activates sporulation in part through different mechanisms from those it uses to activate quiescence [216]. Finally, Rim15 is required for at least some phg-spectrum phenotypes, such as the formation of structured (lacey) colonies [131].
–
PKA also inhibits differentiation through regulators specific to just one or two differentiation fates. For example, PKA phosphorylates and activates the Sok2 transcriptional repressor [217][218]. Active Sok2 directly represses IME1 transcription, hence blocking sporulation [219] Sok2 also indirectly represses FLO11 and other genes required for phg [220][221]. PKA also specifically inhibits spore morphogenesis by inhibiting Smk1 MAPK, which activates this late stage of sporulation [222]. Finally, PKA also specifically inhibits both quiescence and phg by directly (and indirectly) inactivating Yak1 kinase [223][224]. Yak1 is a key activator of proteins required for quiescence [225][226] and phg [227]. Thus, PKA represses all three forms of diploid cell differentiation through an array of general and specific mechanisms.
–
D) TorC1 pathway inhibits sporulation and quiescence/ageing, but activates phg
TorC1 is one of two Tor complexes in yeast with largely separate roles. When nutrients (especially N) are readily available, the TorC1 pathway activates cellular processes needed for rapid growth, such as protein translation. At the same time, this pathway represses processes induced during nutrient limitation such as utilization of poor nitrogen sources, autophagy, and the stress response.
–
Fully activated TorC1 directly represses both sporulation and entry into quiescence. TorC1 represses these programs through activating Sch9 kinase, which in turn inhibits Rim15 activity/nuclear localization [228]. Thus, the TorC11 pathway acts in parallel to PKA in repressing Rim15 (reviewed in [229]). TorC1 also specifically represses sporulation by preventing nuclear localization/activation of Ime1p [172]. Interestingly, a low level of TorC1 is required for IME1 expression; thus sporulation only initiates with moderate TorC1 activity [230]. In addition, once cells become quiescent, activation of TorC1 diminishes their viability (i.e. promotes ageing) [89]. Indeed, calorie restriction slows ageing by decreasing Sch9 activity [229][231][232].
–
In contrast to its role in inhibiting these other differentiation programs, TorC1 stimulates phg through a second branch of the TorC1 pathway, which activates the Tap42 phosphatase [233]. Tap42 in turn stabilizes the Tec1 transcription factor, which binds FLO11 and activates its transcription [234]. Reflecting that optimal phg requires intermediate N concentrations (see Section “Shared environmental cues & distinct fates”), phg (like sporulation) also requires an intermediate level of TorC1 activity [174].
–
E) Snf1 kinase activates all three forms of differentiation
The Crabtree effect depends on the ability of extracellular glucose to repress enzymes required to metabolize other carbon sources. As levels of glucose and other nutrients decline during late stages of growth, repression of Snf1 kinase activity is released. Activated Snf1 in turn triggers the expression and/or activation of enzymes required for respiration and resistance to stress [235][236][237][238], and at the same time stimulates transcription of genes required for differentiation. For example, Snf1 activates FLO11 transcription by phosphorylating Nrg1/Nrg2 transcription factors [167][239][240]. Similarly, Snf1 is required for IME2 transcription even in the absence of respiration [150]. Finally, either hyper-activating or deleting Snf1 shortens chronological lifespan [115][241][242]. Thus, maintaining viability during quiescence may require intermediate levels of Snf1 activity, similar to the intermediate TorC1 activity required for phg and sporulation.
–
F) The Rim101 pathway activates both filamentous and meiotic fates
The Rim101 pathway is primarily activated by alkaline pH (reviewed in [243], and as cells undergo the diauxic shift and extracellular pH increases, this pathway stimulates both pseudohyphal differentiation and sporulation [186]. For example, both FLO11 induction and IME1 induction require Rim101 [182][244][245][246]. Indeed, a RIM101 polymorphism underlies many of the differences between Ʃ1278b and S288C transcriptomes during growth [247].
–
G) Other pathways and pathway interactions
The above signaling pathways are unlikely to be the only ones regulating differentiation. For example, the Rgt2 /glucose induction pathway, which is activated by glucose, represses Ime2 protein stability [248][249], the Hog1 MAPK pathway, which responds to high osmolarity, inhibits pseudohyphal differentiation [250][251], and the cell wall integrity (protein kinase C) pathway, which responds to cell-wall stress, is required both to maintain quiescent cells [252][253] and for pseudohyphal differentiation [254][255][256]. Finally, the Hac1 (unfolded protein response) pathway, which responds to nitrogen starvation, mediates induction of both sporulation and phg [257].
–
Not only does each of the nutrient-signaling pathways in yeast respond to multiple environmental cues, but also these pathways are not insulated from one another. Instead, the PKA, Snf1, TorC1 and other pathways are densely intertwined by virtue of shared components and shared targets (reviewed in [3][61][194]). As just one example, the TorC1 pathway stimulates pseudohyphal differentiation in part by inhibiting Snf1 kinase [258], whereas the Rim101 and Snf1 pathways converge in both inhibiting Nrg1, a transcriptional activator of FLO11 [259]. Thus, nutrient controls on differentiation occur through an intertwined network of pathways.
–
H) Summary: signaling pathways and fate choice
As can be seen from Fig. 4, there is not a Boolean relationship between the four major nutrient signaling pathways and fate choice. In other words, fate choice is not determined by the ON or OFF state of one or more pathways (even relative to a threshold level of activity). Indeed, most of these pathways act in the same way on multiple fates. One exception is the TorC1 pathway, which activates phg while interfering with the other two pathways. However, even TorC1 does not act in a strictly Boolean manner, since several lines of evidence suggest an intermediate level of TorC1 activity, rather than either the fully active or fully inactive state is what activates both phg and sporulation. More generally, because these pathways regulate initiation of all 3 fates, the level of activation of each pathway, rather than a Boolean state, determines fate choice. Further work is required to ask if the activity level of one pathway relative to another contributes to fate choice.
FLEXIBILITY & STABILITY IN DIFFERENTIATION CHOICE
A) Fate choice stability
Another aspect of the relationship between fate choice and environment is the stability and reversibility of this choice. At one level, all three types of differentiated cells are reversible, in the sense that if sufficient nutrients are restored, pseudohyphae, Q cells and asci all re-enter standard cell proliferation. Furthermore, under some circumstances yeast can switch from one fate to another without intervening growth as undifferentiated cells. In this section, the SEDF hypothesis will be considered in the context of the stability /reversibility of fate choice.
–
B) Epigenetic mechanisms stabilize cell fate
Each fate is in part stabilized by genome-wide epigenetic changes. For example, although pseudohyphae can produce ovoid cells when restored to a very nutrient-rich environment, phg is relatively stable to small changes in environment [59][159]. One mechanism for this stability is that the FLO11 promoter is bi-stable, i.e. it stays in either the ON or OFF state for many cell divisions before switching, and this switch depends on chromatin structure and in particular on histone modifying enzymes [260][261] and long non-coding RNA [262][263][264]. FLO11 expression is also stabilized by a prion-like form of the transcription factor, Mot3. Soluble Mot3 represses FL011 transcript, and this repression is released when Mot3 forms heritable prions [265].
–
Both quiescence and sporulation are also characterized by chromatin modifications. For example, genome-wide alterations in histone modification accompany both progression through sporulation [266][267], and transcriptional inactivation after sporulation is complete [268]. Similarly, quiescence is characterized by both global changes in histone modification [78][269], and positioning of RNAPII upstream of many genes poised for induction when nutrients are restored [270].
–
Once cells initiate sporulation, it is only during the early stages of this program that the meiotic fate is directly reversible. Reversibility in this context means that cells in early stages of meiosis can directly re-enter proliferation if resupplied with nutrients (“return to growth”). The loss of this reversibility, termed “commitment to meiosis”, may also have its basis in epigenetics. More specifically, when cells in these early stages are resupplied with growth nutrients, they exit sporulation and resume cell division; however, at about the time of the first meiotic division, cells become irreversibly committed to meiosis, meaning that they complete sporulation even when resupplied with growth nutrients [271][272]. Commitment to meiosis may involve a positive feedback loop regulating transcription of NDT80 [273][274], and as with FLO11, the silenced state of NDT80 requires a histone deacetylase [222][275]. Commitment to meiosis may also involve a second epigenetic mechanism — Rim4 forms an amyloid-like protein that binds transcripts during sporulation to delay their translation until late stages of the program [276], and it may be that binding of the amyloid to these transcripts also protects them from degradation after nutrients are resupplied [277].
–
C) When yeast change their fate
In some circumstances, cells can switch from one differentiation fate to another. For example, the SK1 laboratory strain background can switch from meiotic development to pseudohyphal development in response to a changing environment [143]. Even committed meiotic cells, if arrested at late stages of meiosis, can eventually re-enter the cell division cycle as pseudohyphae [278][279].
–
Even without shifting media or environment, biofilm-like communities of the SK1 strain background form pseudohyphae at their surface that subsequently undergo a further differentiation to form linear asci [56][57]. Similarly, as noted in the Section “Overlapping regulators but distinct fates”, the shared expression pattern between quiescence and sporulation suggest that cells first enter quiescence and then proceed to sporulation, again without being transferred to a new environment. These several lines of evidence indicate that each of three fates can be temporally coordinated with at least one other fate.
–
D) Summary
A variety of epigenetic mechanisms likely provide stability to yeast differentiation fates, but it is interesting that both phg and sporulation fates are stabilized by amyloid/prion proteins as well as by chromatin modification. Despite these mechanisms, differentiating cells can return to undifferentiated proliferation in response to environmental changes – in some cases even before differentiation is complete. Moreover, yeast can switch from one differentiation pathway to another even in the absence of dramatic environmental changes. The reversibility and flexibility of fate choice in yeast is another argument that distinct fates can occur in a similar environment (SEDF), and against a Boolean mechanism of fate choice.
SHARED COMMUNITIES – COORDINATED FATES
A) Introduction
Yeast grow in nature as communities, not as suspended cultures. Given that different fates respond to the same cues and regulators and that these fates sometimes interconvert, one might expect that in the same population and community, some cells might adopt one fate and some cells another. In fact, as described below, this is exactly what happens both in communities and suspended cultures. Thus, the question of whether different fates are triggered by discrete cues and regulators can be asked in the context of yeast populations. As described below, communities that contain cells adapting different fates are particularly relevant to the SEDF hypothesis.
–
B) Fate partitioning within populations
1) Fate choice in cultures. Before discussing fate choice within communities, it is worth considering fate choice in suspended cultures. Microenvironment can vary between different regions of a community, whereas suspended cultures provide the opportunity to measure fate choice within a population with both uniform genotype and uniform environment. One aspect revealed in suspended cultures is the probabilistic nature of fate choice. That is, depending on the conditions of the cultures, a given fraction of the culture adapts a particular fate. As described below, the probabilistic aspect of fate choice is clearest in cultures in which the population in the culture divides into two alternative differentiation fates.
–
Sporulation is particularly useful in comparing cell fates. For most laboratory strains suspended sporulation cultures (e.g. media containing only 2% potassium acetate) contains both sporulated and unsporulated cells. The unsporulated sub-population likely corresponds to Q cells, since this subpopulation can be further subdivided, by cytological markers into viable cells, those undergoing PCD, and those already dead [280].
–
Similar to diploid cultures, haploid suspended cultures grown for extended periods form at least two populations of cells, designated Q and non-quiescent (NQ) cells. Q cells differ from NQ cells in many properties, including density, stress resistance, lifespan, transcriptome and proteome [152][281][282].
–
2) Examples of the spatial organization of cell fate within communities. As they proliferate, yeast naturally form tightly packed multi-cellular communities such as colonies and biofilms (reviewed in [283][284]). Strikingly, differentiation is not homogenously distributed throughout the community, but instead occurs in specific regions. Below are four examples of yeast communities in which cell fate is spatially organized in communities either with respect to undifferentiated cells or with respect to cells adapting a different fate.
–
a) Sporulation patterns in diploid colonies: Patterns of sporulated cells in colonies are easily visible in embedded sections of colonies [182][285]. Specifically, asci are found throughout the top half of mature colonies and also in a thin layer of cells near the agar surface. In contrast, asci are almost completely absent throughout the broad central layer of the colony. Furthermore, the boundaries between sporulating and non-sporulating cells are sharply defined, and this same pattern is observed in a range of laboratory and natural yeast strains. Indeed, strains newly isolated from the wild form this pattern on both FCs and NFCs and on both synthetic and rich media [25].
–
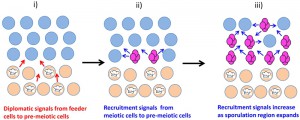
The underlying layer of unsporulated cells can be considered a type of quiescent cell, and are termed “feeder cells” because they remain viable for many days, increase in permeability and stimulate sporulation in the overlying cell layer [74]. As discussed in the Section “Cell-cell signals in communities determine cell fate”, this stimulation is thought to occur because feeder cells provide nutrients to overlying cells, with the increased permeability dependent on induction of the CWI pathway.
–
b) Upper (U) and Lower (L) cell layers in haploid colonies: Haploid colonies form a similar pattern of differentiation as the diploid colonies described above, although of course they do not sporulate. Haploid colonies form two sharply defined layers of cells– termed U and L cells [286][287]. U cells differ from L cells with respect to size, morphology, metabolism, gene expression, and viability.
–
c) Sexual reproduction on the fringe: Both phg and sporulation occur specifically at the edge of some communities. For example, small colonies forming on agar media limiting for nitrogen (microcolonies) form a core of (typically-shaped) ovoid yeast with only the outer surface of the colony containing chains of pseudohyphae projecting from the colony [59]. Similarly, minicolonies, biofilm-like communities growing on plastic surfaces submerged in medium, switch from typical yeast divisions at the early stages of cell growth to phg at the periphery as the growth begins to slow [56]. Biofilms formed by pathogenic yeast such as albicans form a similar pattern in that the underlying layer of cells is comprised of ovoid cells, whereas the top layer consists of hyphae [288][289].
–
As described above, the SK1 strain background both sporulates and undergoes phg very efficiently. As mentioned in the Section “Flexibility & stability in differentiation choice”, the pseudohyphae at the periphery of SK1 microcolonies and minicolonies subsequently sporulate to form linear 2-, 3, and 4-spore asci [56][57]. In minicolonies, the timing of the transition from phg to sporulation has been followed by time-lapse microscopy. In these communities sporulation occurs synchronously around the colony edge but never in the interior of the colony. Furthermore, if phg is blocked, sporulation is also prevented [56]. Thus in these communities, phg and meiotic differentiation are sequential, rather than alternative, fates.
–
d) PCD patterning in colonies: Like sporulation and pseudohyphal differentiation, PCD does not occur uniformly throughout communities. In spot colonies growing on a NFC for 1-3 weeks, apoptosis occurs mostly in the cells that form the core of the colony. In contrast, cells on the rim and outer surface of the colony maintain high viability [290]. It is possible that the high levels of PCD in the core increase the survival or growth among cells at the rim of the same colony, perhaps by providing scarce resources from the PCD region to the viable region [291].
–
3) Sharp boundaries as a common characteristic of community patterning. A striking characteristic of pattern formation in both colonies and biofilms is the sharply defined boundary surrounding regions of differentiation (reviewed in [284]). These boundaries can be visualized not only with respect to cytological markers like apoptosis, spore formation, and phg as described above, but also by patterns of gene expression [74][182][286][287]. Indeed, on one side of a boundary, cells differentiate very efficiently to one fate, whereas neighboring cells in contact with these cells but on the other side of the boundary differentiate equally efficiently but to a different fate.
–
C) Summary
The SEDF hypothesis, that cells can adopt different fates in similar environments, is strongly supported by the sharp boundaries observed between neighboring regions of a yeast community. That is, cells on either side of a boundary share roughly the same environment but adopt distinct fates. In contrast, these boundaries are not easy to reconcile with a Boolean model of fate choice. In the next section we discuss one mechanism allowing SEDF – cell-to-cell signals.
CELL-CELL SIGNALS IN COMMUNITIES DETERMINE CELL FATE
A) Introduction: role of cell-cell signals in cell fate and patterning
As described above, pseudohyphal differentiation, quiescence/aging and sporulation occur under similar conditions (SEDF), are regulated by many of the same signaling pathways and master regulators, and often occur together in the same community. Taken together these results argue against a Boolean relationship between input and cell fate. Given this, how do neighboring cells along a boundary efficiently adopt different fates, and how has yeast evolved to make the correct fate choice across the wide spectrum of environments found in nature? As discussed below, one possible answer to both of these questions is cell-to-cell signaling.
–
Signals between individual yeast cells within communities are typically small diffusible molecules (reviewed in [284]). Below I consider two broad classes of cell-cell signals in yeast: 1) “enlistment” signals, which are produced by differentiating cells within one region of a community to stimulate cells within the same region to adopt the same fate, and 2) “diplomatic signals”, which are produced by differentiating cells in one region of the community to influence cells in an adjoining region to adopt a different fate.
–
B) Enlistment signals—intercellular feedback stimulates differentiation & patterning
Enlistment signals, by reinforcing the same fate choice in neighboring cells, contribute to the spatial organization of communities such that many of the cells within one region of the community adopt the same fate.
–
1) Alkali signals and sporulation patterning. As described in the Section “Shared environmental cues & distinct fates” above, respiration of organic acids during late stages of growth and during sporulation leads to an increase in extracellular pH, which in turn stimulates sporulation in other cells of the same population. In colonies, after a narrow layer of cells near the center initiates sporulation, the resulting alkalization progressively drives expansion of the sporulating region upward to eventually include the entire top half of the colony [182]. This wave of sporulation depends on the Rim101 pathway both to sense and to produce alkali—suggesting an intercellular positive feedback loop involving pH signals.
–
2) Aromatic alcohols and pseudohyphal patterning. Regulation of the dimorphic switch in microcolonies also involves metabolites and an intercellular positive feedback loop [292]. In the case of phg, these signals include the aromatic alcohols phenylethanol and tryptophol. These alcohols are produced and secreted during phg, and in turn stimulate phg in surrounding cells. Moreover, extracellular tryptophol induces further synthesis of tryptophol within pseudohyphal cells, which further amplifies the feedback loop [292].
–
3) Role of ammonium in cycles of proliferation & quiescence. Another example of a cell-cell signal that operates in yeast communities is the ammonium produced by haploid colonies after ≥ 14 d of growth. Cells on the surface of these aged colonies produce ammonium to much higher levels than cells in the core of these colonies [290][293]. At the same time, surface cells induce expression of the ATO1 ammonium exporter [294]. Ammonium contributes to the survival of surface cells and to a new cycle of proliferation at the edge of the colony [295]. Because surface cells both produce and respond to ammonium, ammonium is another example of enlistment signals acting through an intercellular positive feedback loop. Indeed, ammonium can diffuse from one colony to its neighbors, synchronizing these colonies with respect to their growth/quiescence cycles [296].
–
In summary, enlistment signals such as alkali, aromatic alcohols or ammonium may all act through a common mechanism—an intercellular positive-feedback loop. The role of these signals varies; of the three signals discussed above, one activates sporulation, another activates phg, and a third activates proliferation. These feedback loops amplify relatively small differences in nutrient microenvironment to generate larger environmental differences. In this respect, intercellular positive feedback loops contribute to community pattern formation by localizing cell-cell signals to specific regions of a community.
–
C) Diplomatic signals—crossing boundaries to influence behavior
1) The Rlm1 paradigm and the DPEB hypothesis. In contrast to enlistment signals, diplomatic signals occur between community regions adopting different fates. An example of this second type of cell-to-cell signal occurs in sporulating colonies, where activation of the Rlm1 transcription factor in feeder cells stimulates sporulation in an overlying layer of cells through a cell non-autonomous mechanism [74]. Thus, sporulating colonies contain both recruitment and diplomatic signals (Fig. 5).
This type of diplomatic signal may allow yeast colonies to sporulate in a wider range of environments than possible for individual cells, a hypothesis termed, “differential partitioning provides environmental buffering” or DPEB [73][74]. According to this hypothesis, under optimal sporulation conditions, the colony is partitioned such that there are relatively few cells allocated to the feeder cell fate. In contrast, under suboptimal sporulation conditions, a greater portion of the colony is given over to the feeder cell fate, and overall colony sporulation is highly dependent on the nutrients and/or signals provided by these feeder cells. Thus, increasing the proportion of feeder cells buffers sporulation efficiency in suboptimal environments. Differential partitioning is a form of phenotypic plasticity within communities that is related in some ways to task allocation in social organisms such as ant, termite and bee species [297][298].
–
2) Last gasp diplomatic signal. Another example of signals between different regions of the same colony may occur in the haploid colonies undergoing age-triggered PCD described in the Section “Shared communities – coordinated fates”. In these colonies, PCD in the colony core is postulated to provide nutrients to continued cell proliferation at the colony’s rim [290]. Because intracellular ROS levels, including H2O2, which is relatively stable in an extracellular environment, rise in core cells even at relatively early stages of development [299][300], it is possible that peroxide is a diplomatic signal. Thus diplomatic signals, either from feeder cells or PCD cells, may contribute to forming and maintaining the sharp boundaries between regions undergoing alternative fates.
–
D) Summary: Boolean logic and cell-cell signals
In several types of yeast communities, closely neighboring cells on either side of a boundary adopt alternative fates consistent with the SEDF hypothesis but not Boolean models. For yeast to adapt distinct fates in the very similar nutrient environments on either side of a boundary, cell-cell signals are likely essential. The cell-cell signals so far identified may represent only a fraction of the signals operating in yeast communities.
–
Enlistment and diplomatic cell-cell signals cooperate to reinforce small differences in the nutrient microenvironment and are likely important to both establish and maintain boundaries. For example, enlistment signals within one region of a community can amplify differences via intercellular positive feedback loops; whereas, diplomatic signals between neighboring regions can help enforce the sharp boundary between regions. Similar mechanisms are critical to forming boundaries between tissues during metazoan development [301][302]. Finally, cell-cell signals can provide biological function not available to individual cells, such as buffering the efficiency of a differentiation program against unfavorable environments (DPEB).
ORIGINS OF COMMUNICATION & OF MULTICELLULARITY
A) Looking backwards: ancient communities & the birth of communication
What are the broader implications of the non-Boolean SEDF hypothesis and the biological function of pattern formation in yeast communities? Organized patterns within microbial communities date back to the earliest life on earth. For example, stromatolite fossils from billions (109) of years ago provide evidence of organization within cyanobacteria communities (reviewed in [303][304]).
–
The selection pressure for communication to evolve in microbes can be placed in the context of the more complicated forms of communication that evolved in complex organisms. In this respect, it is relevant that some of the most successful metazoan species on earth as judged by total biomass (e.g. humans, termites and ants), are those with highly developed modes of communication. It has been suggested that the “simple multicellularity” that exists in microbial communities differs from the “complex multicellularity” characteristic of plants and animals because of two properties present only in complex organisms: 1) many cells in complex systems do not make contact with the external environment, and 2) high levels of cell-cell communication [305][306]. However, a close look at the biology of yeast communities calls these distinctions into question.
–
B) The advantages of communication & community
Cell signaling between yeast cells hints at the advantages gained from the evolution of communication. In this respect, most of the signaling molecules discussed above share two common characteristics. First, they are important in regulating cell differentiation. Second, they act within the context of a community of yeast, often to partition this community into different regions adopting distinct cell fates.
–
One of the main advantages to yeast of growing within a community may lie in the ability of a community to partition into regions adopting different fates; these regions may cooperate to allow greater biological function than is possible for individual cells. For example, limiting pseudohyphal and meiotic differentiation to the edge of minicolonies and microcolonies maximizes the ability of spores to disperse from the colony [56][57]. As a second example, in colonies the underlying layer of differentiated feeder cells presumably provides nutrients necessary for sporulation in the overlying cell layer, which again is more optimal for spore dispersal. Finally, PCD in the core of a haploid colony may provide the nutrients that allow proliferation at the colony’s rim. Note that because natural yeast communities are largely clonal, the survival of the genotype depends on the overall survival of the colony, rather than competition between S. cerevisiae genotypes in the same community. To extrapolate further, the likely clonal nature of early microbial communities may have led not only to the evolution of the first cell-cell signals on earth but also to the first pattern formation.
–
C) Model for co-evolution of signaling and multicellularity
1) From response to communication. The ability of organisms to respond to their environment is expected even in the earliest life forms. Cell-to-cell signals in modern yeast may yield insight into these ancient signals. In particular, many yeast cell-cell signals are simply metabolites produced by nearby organisms such as alkali generated during respiration or ammonium produced by Q cells. Thus, the earliest form of communication between organisms may have evolved as metabolic byproducts coordinating growth with differentiation.
–
2) From communication to organization. Cell-cell signals in yeast not only regulate cell-fate, they also organize communities such that different regions adopt different fates. By analogy, the response to primordial cell-cell signals may have evolved such that variation in signal concentration across a community contributed to the organization of this community and hence increased fitness.
–
3) From organization to multicellularity. The patterning of cell types in microbial communities may presage cell type patterning within complex multicellular organisms. Indeed, it is conceivable that the earliest multicellular organisms evolved from microbial species with relatively sophisticated cell type patterns. Conversely, patterning in modern microorganisms may provide a useful model for some of the fundamental principles guiding pattern formation in all organisms.
CONCLUSIONS
A) Biology ain’t always Boolean
Similar environmental cues promote each of three alternative differentiation fates (SEDF), and these cues act through many of the same signal transduction pathways and master regulators to control fate choice. Thus, a Boolean representation tracking the presence or absence of a given cue or cues (or a discrete threshold) is not sufficient to describe the relationship between environmental cues and fate choice. Similarly, the relationship between fate choice and signal pathway/master regulator activity also cannot be accurately represented by Boolean logic.
–
B) Graded and specialized responses
So how is fate chosen? In the first place, it is likely that signal transduction pathways are not simply ON or OFF, but have a graded range of activities. By this view, the relative level of activity of signaling pathways determines cells fate as is seen for the TorC1 pathway. For example, the number of activated receptors may vary depending on the concentration of ligand. Indeed, the same signaling pathway might regulate different targets depending on its level of activity. In any case, the activity of a given signal transduction pathway likely also depends on interactions with other signaling pathways.
–
C) Cell-cell signals reinforce differences and provide flexibility
A second aspect to the choice of differentiation fates is provided by cell-cell signals. Relatively small differences in the microenvironment around cells can be reinforced or amplified by both recruitment and diplomatic cell-cell signals. These signals organize yeast communities into cooperative assemblies such that programs such as differentiation or proliferation occur more efficiently than is possible for individual cells. Of particular note is the ability of communities to adjust the allocation of cells to different fates within the community depending on the environment (DPEB). In microorganisms as in more complex organisms, cell-to-cell signals are fundamental to life and may be nearly as ancient.
References
- A. Veliz-Cuba, and B. Stigler, "Boolean Models Can Explain Bistability in the lac Operon", Journal of Computational Biology, vol. 18, pp. 783-794, 2011. http://dx.doi.org/10.1089/cmb.2011.0031
- D. Botstein, and G.R. Fink, "Yeast: An Experimental Organism for 21st Century Biology", Genetics, vol. 189, pp. 695-704, 2011. http://dx.doi.org/10.1534/genetics.111.130765
- S. Zaman, S.I. Lippman, X. Zhao, and J.R. Broach, "HowSaccharomycesResponds to Nutrients", Annual Review of Genetics, vol. 42, pp. 27-81, 2008. http://dx.doi.org/10.1146/annurev.genet.41.110306.130206
- L. Schneper, K. Düvel, and J.R. Broach, "Sense and sensibility: nutritional response and signal integration in yeast", Current Opinion in Microbiology, vol. 7, pp. 624-630, 2004. http://dx.doi.org/10.1016/j.mib.2004.10.002
- A.M. Neiman, "Sporulation in the Budding Yeast Saccharomyces cerevisiae", Genetics, vol. 189, pp. 737-765, 2011. http://dx.doi.org/10.1534/genetics.111.127126
- M. Kupiec, B. Byers, R. Esposito, and A. Mitchell, "Meiosis and sporulation in Saccharomyces cerevisiae. In: Pringle JR, Broach JR, Jones EW, editors. The molecular and cellular biology of the yeast Saccharomyces: Cell cycle and cell biology.", Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY; pp 889-1036., 1997.
- J.R. Dickinson, "Filament formation in Saccharomyces cerevisiae — a review", Folia Microbiologica, vol. 53, pp. 3-14, 2008. http://dx.doi.org/10.1007/s12223-008-0001-6
- M. Gagiano, F.F. Bauer, and I.S. Pretorius, "The sensing of nutritional status and the relationship to filamentous growth inSaccharomyces cerevisiae", FEMS Yeast Research, vol. 2, pp. 433-470, 2002. http://dx.doi.org/10.1111/j.1567-1364.2002.tb00114.x
- J.M. Gancedo, "Control of pseudohyphae formation inSaccharomyces cerevisiae", FEMS Microbiology Reviews, vol. 25, pp. 107-123, 2001. http://dx.doi.org/10.1111/j.1574-6976.2001.tb00573.x
- Z. Palková, D. Wilkinson, and L. Váchová, "Aging and differentiation in yeast populations: elders with different properties and functions", FEMS Yeast Research, vol. 14, pp. 96-108, 2013. http://dx.doi.org/10.1111/1567-1364.12103
- C. De Virgilio, "The essence of yeast quiescence", FEMS Microbiology Reviews, vol. 36, pp. 306-339, 2012. http://dx.doi.org/10.1111/j.1574-6976.2011.00287.x
- J.V. Gray, G.A. Petsko, G.C. Johnston, D. Ringe, R.A. Singer, and M. Werner-Washburne, "“Sleeping Beauty”: Quiescence inSaccharomyces cerevisiae", Microbiology and Molecular Biology Reviews, vol. 68, pp. 187-206, 2004. http://dx.doi.org/10.1128/MMBR.68.2.187-206.2004
- G. Liti, D.M. Carter, A.M. Moses, J. Warringer, L. Parts, S.A. James, R.P. Davey, I.N. Roberts, A. Burt, V. Koufopanou, I.J. Tsai, C.M. Bergman, D. Bensasson, M.J.T. O’Kelly, A. van Oudenaarden, D.B.H. Barton, E. Bailes, A.N. Nguyen, M. Jones, M.A. Quail, I. Goodhead, S. Sims, F. Smith, A. Blomberg, R. Durbin, and E.J. Louis, "Population genomics of domestic and wild yeasts", Nature, vol. 458, pp. 337-341, 2009. http://dx.doi.org/10.1038/nature07743
- G.I. Naumov, E.S. Naumova, and P.D. Sniegowski, "Saccharomyces paradoxus and Saccharomyces cerevisiae are associated with exudates of North American oaks", Canadian Journal of Microbiology, vol. 44, pp. 1045-1050, 1998. http://dx.doi.org/10.1139/w98-104
-
Q. Wang, W. Liu, G. Liti, S. Wang, and F. Bai, "Surprisingly diverged populations of
S accharomyces cerevisiae in natural environments remote from human activity", Molecular Ecology, vol. 21, pp. 5404-5417, 2012. http://dx.doi.org/10.1111/j.1365-294X.2012.05732.x - E. Aa, J.P. Townsend, R.I. Adams, K.M. Nielsen, and J.W. Taylor, "Population structure and gene evolution inSaccharomyces cerevisiae", FEMS Yeast Research, vol. 6, pp. 702-715, 2006. http://dx.doi.org/10.1111/j.1567-1364.2006.00059.x
- J.C. Fay, and J.A. Benavides, "Evidence for Domesticated and Wild Populations of Saccharomyces cerevisiae", PLoS Genetics, vol. 1, pp. e5, 2005. http://dx.doi.org/10.1371/journal.pgen.0010005
- J. Hose, C.M. Yong, M. Sardi, Z. Wang, M.A. Newton, and A.P. Gasch, "Dosage compensation can buffer copy-number variation in wild yeast", eLife, vol. 4, 2015. http://dx.doi.org/10.7554/eLife.05462
- F.A. Cubillos, E.J. Louis, and G. Liti, "Generation of a large set of genetically tractable haploid and diploidSaccharomycesâstrains", FEMS Yeast Research, vol. 9, pp. 1217-1225, 2009. http://dx.doi.org/10.1111/j.1567-1364.2009.00583.x
- E.A. Hope, and M.J. Dunham, "Ploidy-Regulated Variation in Biofilm-Related Phenotypes in Natural Isolates of Saccharomyces cerevisiae", G3 Genes|Genomes|Genetics, vol. 4, pp. 1773-1786, 2014. http://dx.doi.org/10.1534/g3.114.013250
- W. Albertin, and P. Marullo, "Polyploidy in fungi: evolution after whole-genome duplication", Proceedings of the Royal Society B: Biological Sciences, vol. 279, pp. 2497-2509, 2012. http://dx.doi.org/10.1098/rspb.2012.0434
- D. Sicard, and J. Legras, "Bread, beer and wine: Yeast domestication in the Saccharomyces sensu stricto complex", Comptes Rendus. Biologies, vol. 334, pp. 229-236, 2011. http://dx.doi.org/10.1016/j.crvi.2010.12.016
- Z. Storchova, "Ploidy changes and genome stability in yeast", Yeast, vol. 31, pp. 421-430, 2014. http://dx.doi.org/10.1002/yea.3037
- J.P. Gerke, C.T.L. Chen, and B.A. Cohen, "Natural Isolates of Saccharomyces cerevisiae Display Complex Genetic Variation in Sporulation Efficiency", Genetics, vol. 174, pp. 985-997, 2006. http://dx.doi.org/10.1534/genetics.106.058453
- S. Piccirillo, and S.M. Honigberg, "Sporulation patterning and invasive growth in wild and domesticated yeast colonies", Research in Microbiology, vol. 161, pp. 390-398, 2010. http://dx.doi.org/10.1016/j.resmic.2010.04.001
- P.D. Sniegowski, P.G. Dombrowski, and E. Fingerman, "Saccharomyces cerevisiaeandSaccharomyces paradoxuscoexist in a natural woodland site in North America and display different levels of reproductive isolation from European conspecifics", FEMS Yeast Research, vol. 1, pp. 299-306, 2002. http://dx.doi.org/10.1111/j.1567-1364.2002.tb00048.x
- E. Zörgö, K. Chwialkowska, A.B. Gjuvsland, E. Garré, P. Sunnerhagen, G. Liti, A. Blomberg, S.W. Omholt, and J. Warringer, "Ancient Evolutionary Trade-Offs between Yeast Ploidy States", PLoS Genetics, vol. 9, pp. e1003388, 2013. http://dx.doi.org/10.1371/journal.pgen.1003388
- R.K. Sherwood, C.M. Scaduto, S.E. Torres, and R.J. Bennett, "Convergent evolution of a fused sexual cycle promotes the haploid lifestyle", Nature, vol. 506, pp. 387-390, 2014. http://dx.doi.org/10.1038/nature12891
- L. Merlini, O. Dudin, and S.G. Martin, "Mate and fuse: how yeast cells do it", Open Biology, vol. 3, pp. 130008, 2013. http://dx.doi.org/10.1098/rsob.130008
- D.M. Ruderfer, S.C. Pratt, H.S. Seidel, and L. Kruglyak, "Population genomic analysis of outcrossing and recombination in yeast", Nature Genetics, vol. 38, pp. 1077-1081, 2006. http://dx.doi.org/10.1038/ng1859
- S. Diezmann, and F.S. Dietrich, "Saccharomyces cerevisiae: Population Divergence and Resistance to Oxidative Stress in Clinical, Domesticated and Wild Isolates", PLoS ONE, vol. 4, pp. e5317, 2009. http://dx.doi.org/10.1371/journal.pone.0005317
- P.M. Magwene, �. Kayıkçı, J.A. Granek, J.M. Reininga, Z. Scholl, and D. Murray, "Outcrossing, mitotic recombination, and life-history trade-offs shape genome evolution in Saccharomyces cerevisiae", Proceedings of the National Academy of Sciences, vol. 108, pp. 1987-1992, 2011. http://dx.doi.org/10.1073/pnas.1012544108
- I. Stefanini, L. Dapporto, L. Berná, M. Polsinelli, S. Turillazzi, and D. Cavalieri, "Social wasps are a Saccharomyces mating nest", Proceedings of the National Academy of Sciences, vol. 113, pp. 2247-2251, 2016. http://dx.doi.org/10.1073/pnas.1516453113
- M. Knop, "Evolution of the hemiascomycete yeasts: on life styles and the importance of inbreeding", BioEssays, vol. 28, pp. 696-708, 2006. http://dx.doi.org/10.1002/bies.20435
- A.P. Mitchell, and I. Herskowitz, "Activation of meiosis and sporulation by repression of the RME1 product in yeast", Nature, vol. 319, pp. 738-742, 1986. http://dx.doi.org/10.1038/319738a0
- J.E. Wagstaff, S. Klapholz, and R.E. Esposito, "Meiosis in haploid yeast", Proceedings of the National Academy of Sciences, vol. 79, pp. 2986-2990, 1982. http://dx.doi.org/10.1073/pnas.79.9.2986
- J.P. van Dijken, R.A. Weusthuis, and J.T. Pronk, "Kinetics of growth and sugar consumption in yeasts.", Antonie van Leeuwenhoek, 1993. http://www.ncbi.nlm.nih.gov/pubmed/8279829
- J. Kim, A. Roy, D. Jouandot, and K.H. Cho, "The glucose signaling network in yeast", Biochimica et Biophysica Acta (BBA) - General Subjects, vol. 1830, pp. 5204-5210, 2013. http://dx.doi.org/10.1016/j.bbagen.2013.07.025
- S. Dashko, N. Zhou, C. Compagno, and J. Piškur, "Why, when, and how did yeast evolve alcoholic fermentation?", FEMS Yeast Research, vol. 14, pp. 826-832, 2014. http://dx.doi.org/10.1111/1567-1364.12161
- M.R. Goddard, and D. Greig, "Saccharomyces cerevisiae: a nomadic yeast with no niche?", FEMS Yeast Research, vol. 15, 2015. http://dx.doi.org/10.1093/femsyr/fov009
- N. Paczia, A. Nilgen, T. Lehmann, J. Gätgens, W. Wiechert, and S. Noack, "Extensive exometabolome analysis reveals extended overflow metabolism in various microorganisms", Microbial Cell Factories, vol. 11, 2012. http://dx.doi.org/10.1186/1475-2859-11-122
- G.G. Zampar, A. Kümmel, J. Ewald, S. Jol, B. Niebel, P. Picotti, R. Aebersold, U. Sauer, N. Zamboni, and M. Heinemann, "Temporal system‐level organization of the switch from glycolytic to gluconeogenic operation in yeast", Molecular Systems Biology, vol. 9, 2013. http://dx.doi.org/10.1038/msb.2013.11
- H. Schüller, "Transcriptional control of nonfermentative metabolism in the yeast Saccharomyces cerevisiae.", Current genetics, 2003. http://www.ncbi.nlm.nih.gov/pubmed/12715202
- R. Kumar, S. Dhali, R. Srikanth, S.K. Ghosh, and S. Srivastava, "Comparative proteomics of mitosis and meiosis in Saccharomyces cerevisiae", Journal of Proteomics, vol. 109, pp. 1-15, 2014. http://dx.doi.org/10.1016/j.jprot.2014.06.006
- J. Grassl, C. Scaife, J. Polden, C.N. Daly, M.G. Iacovella, M.J. Dunn, and R.K. Clyne, "Analysis of the budding yeast pH 4–7 proteome in meiosis", PROTEOMICS, vol. 10, pp. 506-519, 2010. http://dx.doi.org/10.1002/pmic.200900561
- T. Walther, F. Létisse, L. Peyriga, C. Alkim, Y. Liu, A. Lardenois, H. Martin-Yken, J. Portais, M. Primig, and J.M. François\, "Developmental stage-dependent metabolic regulation during meiotic differentiation in budding yeast", BMC Biology, vol. 12, 2014. http://dx.doi.org/10.1186/s12915-014-0060-x
- D. Ray, and P. Ye, "Characterization of the Metabolic Requirements in Yeast Meiosis", PLoS ONE, vol. 8, pp. e63707, 2013. http://dx.doi.org/10.1371/journal.pone.0063707
- R.K. Mortimer, and J.R. Johnston, "Genealogy of principal strains of the yeast genetic stock center.", Genetics, 1986. http://www.ncbi.nlm.nih.gov/pubmed/3519363
- A.M. Deutschbauer, and R.W. Davis, "Quantitative trait loci mapped to single-nucleotide resolution in yeast", Nature Genetics, vol. 37, pp. 1333-1340, 2005. http://dx.doi.org/10.1038/ng1674
- G. Ben-Ari, D. Zenvirth, A. Sherman, L. David, M. Klutstein, U. Lavi, J. Hillel, and G. Simchen, "Four Linked Genes Participate in Controlling Sporulation Efficiency in Budding Yeast", PLoS Genetics, vol. 2, pp. e195, 2006. http://dx.doi.org/10.1371/journal.pgen.0020195
- E. Casalone, C. Barberio, L. Cappellini, and M. Polsinelli, "Characterization of Saccharomyces cerevisiae natural populations for pseudohyphal growth and colony morphology", Research in Microbiology, vol. 156, pp. 191-200, 2005. http://dx.doi.org/10.1016/j.resmic.2004.09.008
- Q. Song, C. Johnson, T.E. Wilson, and A. Kumar, "Pooled Segregant Sequencing Reveals Genetic Determinants of Yeast Pseudohyphal Growth", PLoS Genetics, vol. 10, pp. e1004570, 2014. http://dx.doi.org/10.1371/journal.pgen.1004570
- H. Liu, C.A. Styles, and G.R. Fink, "Saccharomyces cerevisiae S288C has a mutation in FLO8, a gene required for filamentous growth.", Genetics, 1996. http://www.ncbi.nlm.nih.gov/pubmed/8913742
- S. Jubany, I. Tomasco, I. Ponce de León, K. Medina, F. Carrau, N. Arrambide, H. Naya, and C. Gaggero, "Toward a global database for the molecular typing of Saccharomyces cerevisiae strains", FEMS Yeast Research, vol. 8, pp. 472-484, 2008. http://dx.doi.org/10.1111/j.1567-1364.2008.00361.x
- S.E. Van Mulders, E. Christianen, S.M. Saerens, L. Daenen, P.J. Verbelen, R. Willaert, K.J. Verstrepen, and F.R. Delvaux, "Phenotypic diversity of Flo protein family-mediated adhesion inSaccharomyces cerevisiae", FEMS Yeast Research, vol. 9, pp. 178-190, 2009. http://dx.doi.org/10.1111/j.1567-1364.2008.00462.x
- M.G. White, S. Piccirillo, V. Dusevich, D.J. Law, T. Kapros, and S.M. Honigberg, "Flo11p adhesin required for meiotic differentiation in Saccharomyces cerevisiae minicolonies grown on plastic surfaces", FEMS Yeast Research, vol. 11, pp. 223-232, 2011. http://dx.doi.org/10.1111/j.1567-1364.2010.00712.x
- N. Strudwick, M. Brown, V.M. Parmar, and M. Schröder, "Ime1 and Ime2 Are Required for Pseudohyphal Growth of Saccharomyces cerevisiae on Nonfermentable Carbon Sources", Molecular and Cellular Biology, vol. 30, pp. 5514-5530, 2010. http://dx.doi.org/10.1128/MCB.00390-10
- J. BAYLY, L. DOUGLAS, I. PRETORIUS, F. BAUER, and A. DRANGINIS, "Characteristics of Flo11-dependent flocculation in", FEMS Yeast Research, vol. 5, pp. 1151-1156, 2005. http://dx.doi.org/10.1016/j.femsyr.2005.05.004
- C.J. Gimeno, P.O. Ljungdahl, C.A. Styles, and G.R. Fink, "Unipolar cell divisions in the yeast S. cerevisiae lead to filamentous growth: Regulation by starvation and RAS", Cell, vol. 68, pp. 1077-1090, 1992. http://dx.doi.org/10.1016/0092-8674(92)90079-r
- P. Fabrizio, C. Gattazzo, L. Battistella, M. Wei, C. Cheng, K. McGrew, and V.D. Longo, "Sir2 Blocks Extreme Life-Span Extension", Cell, vol. 123, pp. 655-667, 2005. http://dx.doi.org/10.1016/j.cell.2005.08.042
- D. Engelberg, R. Perlman, and A. Levitzki, "Transmembrane signaling in Saccharomyces cerevisiae as a model for signaling in metazoans: State of the art after 25years", Cellular Signalling, vol. 26, pp. 2865-2878, 2014. http://dx.doi.org/10.1016/j.cellsig.2014.09.003
- A.E. Coluccio, R.K. Rodriguez, M.J. Kernan, and A.M. Neiman, "The Yeast Spore Wall Enables Spores to Survive Passage through the Digestive Tract of Drosophila", PLoS ONE, vol. 3, pp. e2873, 2008. http://dx.doi.org/10.1371/journal.pone.0002873
- D. Granot, and M. Snyder, "Glucose induces cAMP-independent growth-related changes in stationary-phase cells of Saccharomyces cerevisiae.", Proceedings of the National Academy of Sciences, vol. 88, pp. 5724-5728, 1991. http://dx.doi.org/10.1073/pnas.88.13.5724
- P. Romano, and G. Suzzi, "Sensitivity of Saccharomyces cerevisiae vegetative cells and spores to antimicrobial compounds", Journal of Applied Bacteriology, vol. 59, pp. 299-302, 1985. http://dx.doi.org/10.1111/j.1365-2672.1985.tb03323.x
- P. Briza, M. Breitenbach, A. Ellinger, and J. Segall, "Isolation of two developmentally regulated genes involved in spore wall maturation in Saccharomyces cerevisiae.", Genes & Development, vol. 4, pp. 1775-1789, 1990. http://dx.doi.org/10.1101/gad.4.10.1775
- G. Muthukumar, S.H. Suhng, P.T. Magee, R.D. Jewell, and D.A. Primerano, "The Saccharomyces cerevisiae SPR1 gene encodes a sporulation-specific exo-1,3-beta-glucanase which contributes to ascospore thermoresistance.", Journal of bacteriology, 1993. http://www.ncbi.nlm.nih.gov/pubmed/8419289
- M. Reuter, G. Bell, and D. Greig, "Increased outbreeding in yeast in response to dispersal by an insect vector", Current Biology, vol. 17, pp. R81-R83, 2007. http://dx.doi.org/10.1016/j.cub.2006.11.059
- M. Lachance, D.G. Gilbert, and W.T. Starmer, "Yeast communities associated withDrosophila species and related flies in an eastern oak-pine forest: A comparison with western communities", Journal of Industrial Microbiology, vol. 14, pp. 484-494, 1995. http://dx.doi.org/10.1007/bf01573963
- A. Rattray, G. Santoyo, B. Shafer, and J.N. Strathern, "Elevated Mutation Rate during Meiosis in Saccharomyces cerevisiae", PLoS Genetics, vol. 11, pp. e1004910, 2015. http://dx.doi.org/10.1371/journal.pgen.1004910
- M. Primig, R.M. Williams, E.A. Winzeler, G.G. Tevzadze, A.R. Conway, S.Y. Hwang, R.W. Davis, and R.E. Esposito, "The core meiotic transcriptome in budding yeasts", Nature Genetics, vol. 26, pp. 415-423, 2000. http://dx.doi.org/10.1038/82539
- S. Chu, J. DeRisi, M. Eisen, J. Mulholland, D. Botstein, P.O. Brown, and I. Herskowitz, "The Transcriptional Program of Sporulation in Budding Yeast", Science, vol. 282, pp. 699-705, 1998. http://dx.doi.org/10.1126/science.282.5389.699
- E. Becker, Y. Liu, A. Lardenois, T. Walther, J. Horecka, I. Stuparevic, M.J. Law, R. Lavigne, B. Evrard, P. Demougin, M. Riffle, R. Strich, R.W. Davis, C. Pineau, and M. Primig, "Integrated RNA- and protein profiling of fermentation and respiration in diploid budding yeast provides insight into nutrient control of cell growth and development", Journal of Proteomics, vol. 119, pp. 30-44, 2015. http://dx.doi.org/10.1016/j.jprot.2015.01.015
- S. Piccirillo, T. Kapros, and S.M. Honigberg, "Phenotypic plasticity within yeast colonies: differential partitioning of cell fates", Current Genetics, vol. 62, pp. 467-473, 2016. http://dx.doi.org/10.1007/s00294-015-0558-y
- S. Piccirillo, R. Morales, M.G. White, K. Smith, T. Kapros, and S.M. Honigberg, "Cell Differentiation and Spatial Organization in Yeast Colonies: Role of Cell-Wall Integrity Pathway", Genetics, vol. 201, pp. 1427-1438, 2015. http://dx.doi.org/10.1534/genetics.115.180919
- R. Esposito, and S. Klapholz, "Meiosis and ascospore development. In: Strathern JN, Jones EW, Broach JR, editors. Molecular biology of the yeast Saccharomyces: Life cycle and inheritance.", Cold Spring Harbor Laboratory, Cold Spring Harbor, NY; pp 211-287., 1981.
- M.N. De Silva‐Udawatta, and J.F. Cannon, "Roles of trehalose phosphate synthase in yeast glycogen metabolism and sporulation", Molecular Microbiology, vol. 40, pp. 1345-1356, 2001. http://dx.doi.org/10.1046/j.1365-2958.2001.02477.x
- N.T. Ramaswamy, L. Li, M. Khalil, and J.F. Cannon, "Regulation of yeast glycogen metabolism and sporulation by Glc7p protein phosphatase.", Genetics, 1998. http://www.ncbi.nlm.nih.gov/pubmed/9584086
- J. McKnight, J. Boerma, L. Breeden, and T. Tsukiyama, "Global Promoter Targeting of a Conserved Lysine Deacetylase for Transcriptional Shutoff during Quiescence Entry", Molecular Cell, vol. 59, pp. 732-743, 2015. http://dx.doi.org/10.1016/j.molcel.2015.07.014
- S. Miles, L. Li, J. Davison, and L.L. Breeden, "Xbp1 Directs Global Repression of Budding Yeast Transcription during the Transition to Quiescence and Is Important for the Longevity and Reversibility of the Quiescent State", PLoS Genetics, vol. 9, pp. e1003854, 2013. http://dx.doi.org/10.1371/journal.pgen.1003854
- M.T. Rutledge, M. Russo, J. Belton, J. Dekker, and J.R. Broach, "The yeast genome undergoes significant topological reorganization in quiescence", Nucleic Acids Research, vol. 43, pp. 8299-8313, 2015. http://dx.doi.org/10.1093/nar/gkv723
- D. Laporte, F. Courtout, B. Salin, J. Ceschin, and I. Sagot, "An array of nuclear microtubules reorganizes the budding yeast nucleus during quiescence", Journal of Cell Biology, vol. 203, pp. 585-594, 2013. http://dx.doi.org/10.1083/jcb.201306075
- M.C. Munder, D. Midtvedt, T. Franzmann, E. Nüske, O. Otto, M. Herbig, E. Ulbricht, P. Müller, A. Taubenberger, S. Maharana, L. Malinovska, D. Richter, J. Guck, V. Zaburdaev, and S. Alberti, "A pH-driven transition of the cytoplasm from a fluid- to a solid-like state promotes entry into dormancy", eLife, vol. 5, 2016. http://dx.doi.org/10.7554/eLife.09347
- R.P. Joyner, J.H. Tang, J. Helenius, E. Dultz, C. Brune, L.J. Holt, S. Huet, D.J. Müller, and K. Weis, "A glucose-starvation response regulates the diffusion of macromolecules", eLife, vol. 5, 2016. http://dx.doi.org/10.7554/eLife.09376
- L. Li, S. Miles, and L.L. Breeden, "A Genetic Screen forSaccharomyces cerevisiaeMutants That Fail to Enter Quiescence", G3 Genes|Genomes|Genetics, vol. 5, pp. 1783-1795, 2015. http://dx.doi.org/10.1534/g3.115.019091
- J. Torres, C.J. Di Como, E. Herrero, and M.A. de la Torre-Ruiz, "Regulation of the Cell Integrity Pathway by Rapamycin-sensitive TOR Function in Budding Yeast", Journal of Biological Chemistry, vol. 277, pp. 43495-43504, 2002. http://dx.doi.org/10.1074/jbc.M205408200
- E. Cameroni, N. Hulo, J. Roosen, J. Winderickx, and C.D. Virgilio, "The Novel Yeast PAS Kinase Rim15 Orchestrates G0-Associated Antioxidant Defense Mechanisms", Cell Cycle, vol. 3, pp. 460-466, 2004. http://dx.doi.org/10.4161/cc.3.4.791
- N. Zhang, J. Wu, and S.G. Oliver, "Gis1 is required for transcriptional reprogramming of carbon metabolism and the stress response during transition into stationary phase in yeast", Microbiology, vol. 155, pp. 1690-1698, 2009. http://dx.doi.org/10.1099/mic.0.026377-0
- J. Roosen, K. Engelen, K. Marchal, J. Mathys, G. Griffioen, E. Cameroni, J.M. Thevelein, C. De Virgilio, B. De Moor, and J. Winderickx, "PKA and Sch9 control a molecular switch important for the proper adaptation to nutrient availability", Molecular Microbiology, vol. 55, pp. 862-880, 2004. http://dx.doi.org/10.1111/j.1365-2958.2004.04429.x
- R.W. Powers, M. Kaeberlein, S.D. Caldwell, B.K. Kennedy, and S. Fields, "Extension of chronological life span in yeast by decreased TOR pathway signaling", Genes & Development, vol. 20, pp. 174-184, 2006. http://dx.doi.org/10.1101/gad.1381406
- P. Fabrizio, F. Pozza, S.D. Pletcher, C.M. Gendron, and V.D. Longo, "Regulation of Longevity and Stress Resistance by Sch9 in Yeast", Science, vol. 292, pp. 288-290, 2001. http://dx.doi.org/10.1126/science.1059497
- D. Carmona‐Gutierrez, and S. Büttner, "The many ways to age for a single yeast cell", Yeast, vol. 31, pp. 289-298, 2014. http://dx.doi.org/10.1002/yea.3020
- C. Ruckenstuhl, D. Carmona-Gutierrez, and F. Madeo, "The sweet taste of death: glucose triggers apoptosis during yeast chronological aging", Aging, vol. 2, pp. 643-649, 2010. http://dx.doi.org/10.18632/aging.100223
- P. Fabrizio, and V.D. Longo, "Chronological aging-induced apoptosis in yeast", Biochimica et Biophysica Acta (BBA) - Molecular Cell Research, vol. 1783, pp. 1280-1285, 2008. http://dx.doi.org/10.1016/j.bbamcr.2008.03.017
- D. Wloch-Salamon, and A. Bem, "Types of cell death and methods of their detection in yeast Saccharomyces cerevisiae", Journal of Applied Microbiology, vol. 114, pp. 287-298, 2012. http://dx.doi.org/10.1111/jam.12024
- R. Strich, "Programmed Cell Death Initiation and Execution in Budding Yeast", Genetics, vol. 200, pp. 1003-1014, 2015. http://dx.doi.org/10.1534/genetics.115.179150
- M.T. Greenwood, and P. Ludovico, "Expressing and functional analysis of mammalian apoptotic regulators in yeast", Cell Death & Differentiation, vol. 17, pp. 737-745, 2009. http://dx.doi.org/10.1038/cdd.2009.177
- L. Váchová, and Z. Palková, "Caspases in yeast apoptosis-like death: facts and artefacts", FEMS Yeast Research, vol. 7, pp. 12-21, 2007. http://dx.doi.org/10.1111/j.1567-1364.2006.00137.x
- D. Sinclair, K. Mills, and L. Guarente, "AGING IN SACCHAROMYCES CEREVISIAE", Annual Review of Microbiology, vol. 52, pp. 533-560, 1998. http://dx.doi.org/10.1146/annurev.micro.52.1.533
- V. Longo, G. Shadel, M. Kaeberlein, and B. Kennedy, "Replicative and Chronological Aging in Saccharomyces cerevisiae", Cell Metabolism, vol. 16, pp. 18-31, 2012. http://dx.doi.org/10.1016/j.cmet.2012.06.002
- M. Kaeberlein, M. McVey, and L. Guarente, "Using Yeast to Discover the Fountain of Youth", Science of Aging Knowledge Environment, vol. 2001, 2001. http://dx.doi.org/10.1126/sageke.2001.1.pe1
- K. Ashrafi, D. Sinclair, J.I. Gordon, and L. Guarente, "Passage through stationary phase advances replicative aging in Saccharomyces cerevisiae", Proceedings of the National Academy of Sciences, vol. 96, pp. 9100-9105, 1999. http://dx.doi.org/10.1073/pnas.96.16.9100
- M.B. Wierman, and J.S. Smith, "Yeast sirtuins and the regulation of aging", FEMS Yeast Research, vol. 14, pp. 73-88, 2013. http://dx.doi.org/10.1111/1567-1364.12115
- Q. Gao, Q. Ren, L. Liou, X. Bao, and Z. Zhang, "Mitochondrial DNA protects against salt stress‐induced cytochrome c‐mediated apoptosis in yeast", FEBS Letters, vol. 585, pp. 2507-2512, 2011. http://dx.doi.org/10.1016/j.febslet.2011.06.034
- K. Sapienza, W. Bannister, and R. Balzan, "Mitochondrial involvement in aspirin-induced apoptosis in yeast", Microbiology, vol. 154, pp. 2740-2747, 2008. http://dx.doi.org/10.1099/mic.0.2008/017228-0
- P. Ludovico, M.J. Sousa, M.T. Silva, C. Leão, and M. Côrte-Real, "Saccharomyces cerevisiae commits to a programmed cell death process in response to acetic acid", Microbiology, vol. 147, pp. 2409-2415, 2001. http://dx.doi.org/10.1099/00221287-147-9-2409
- F. Madeo, E. Fröhlich, M. Ligr, M. Grey, S.J. Sigrist, D.H. Wolf, and K. Fröhlich, "Oxygen Stress: A Regulator of Apoptosis in Yeast", The Journal of Cell Biology, vol. 145, pp. 757-767, 1999. http://dx.doi.org/10.1083/jcb.145.4.757
- V.D. Longo, and P. Fabrizio, "Chronological Aging in Saccharomyces cerevisiae", Aging Research in Yeast, pp. 101-121, 2011. http://dx.doi.org/10.1007/978-94-007-2561-4_5
- N.A. Bishop, and L. Guarente, "Genetic links between diet and lifespan: shared mechanisms from yeast to humans", Nature Reviews Genetics, vol. 8, pp. 835-844, 2007. http://dx.doi.org/10.1038/nrg2188
- S. Lin, P. Defossez, and L. Guarente, "Requirement of NAD and SIR2 for Life-Span Extension by Calorie Restriction in Saccharomyces cerevisiae", Science, vol. 289, pp. 2126-2128, 2000. http://dx.doi.org/10.1126/science.289.5487.2126
- A. Ayer, C.W. Gourlay, and I.W. Dawes, "Cellular redox homeostasis, reactive oxygen species and replicative ageing inSaccharomyces cerevisiae", FEMS Yeast Research, vol. 14, pp. 60-72, 2013. http://dx.doi.org/10.1111/1567-1364.12114
- T. Finkel, and N.J. Holbrook, "Oxidants, oxidative stress and the biology of ageing", Nature, vol. 408, pp. 239-247, 2000. http://dx.doi.org/10.1038/35041687
- R.S. Sohal, and R. Weindruch, "Oxidative Stress, Caloric Restriction, and Aging", Science, vol. 273, pp. 59-63, 1996. http://dx.doi.org/10.1126/science.273.5271.59
- J. Ring, C. Sommer, D. Carmona-Gutierrez, C. Ruckenstuhl, T. Eisenberg, and F. Madeo, "The metabolism beyond programmed cell death in yeast", Experimental Cell Research, vol. 318, pp. 1193-1200, 2012. http://dx.doi.org/10.1016/j.yexcr.2012.03.019
- A. Ocampo, J. Liu, E. Schroeder, G. Shadel, and A. Barrientos, "Mitochondrial Respiratory Thresholds Regulate Yeast Chronological Life Span and its Extension by Caloric Restriction", Cell Metabolism, vol. 16, pp. 55-67, 2012. http://dx.doi.org/10.1016/j.cmet.2012.05.013
- M. Weinberger, A. Mesquita, T. Carroll, L. Marks, H. Yang, Z. Zhang, P. Ludovico, and W.C. Burhans, "Growth signaling promotes chronological aging in budding yeast by inducing superoxide anions that inhibit quiescence", Aging, vol. 2, pp. 709-726, 2010. http://dx.doi.org/10.18632/aging.100215
- P. Fabrizio, L. Battistella, R. Vardavas, C. Gattazzo, L. Liou, A. Diaspro, J.W. Dossen, E.B. Gralla, and V.D. Longo, "Superoxide is a mediator of an altruistic aging program in Saccharomyces cerevisiae ", The Journal of Cell Biology, vol. 166, pp. 1055-1067, 2004. http://dx.doi.org/10.1083/jcb.200404002
- F.F. Severin, M.V. Meer, E.A. Smirnova, D.A. Knorre, and V.P. Skulachev, "Natural causes of programmed death of yeast Saccharomyces cerevisiae", Biochimica et Biophysica Acta (BBA) - Molecular Cell Research, vol. 1783, pp. 1350-1353, 2008. http://dx.doi.org/10.1016/j.bbamcr.2008.02.001
- N. Allocati, M. Masulli, C. Di Ilio, and V. De Laurenzi, "Die for the community: an overview of programmed cell death in bacteria", Cell Death & Disease, vol. 6, pp. e1609-e1609, 2015. http://dx.doi.org/10.1038/cddis.2014.570
- K. Lewis, "Programmed Death in Bacteria", Microbiology and Molecular Biology Reviews, vol. 64, pp. 503-514, 2000. http://dx.doi.org/10.1128/mmbr.64.3.503-514.2000
- J.C. Ameisen, "The Origin of Programmed Cell Death", Science, vol. 272, pp. 1278-1279, 1996. http://dx.doi.org/10.1126/science.272.5266.1278
- S.J. Kron, C.A. Styles, and G.R. Fink, "Symmetric cell division in pseudohyphae of the yeast Saccharomyces cerevisiae.", Molecular Biology of the Cell, vol. 5, pp. 1003-1022, 1994. http://dx.doi.org/10.1091/mbc.5.9.1003
- J. Dickinson, "Irreversible formation of pseudohyphae by haploidSaccharomyces cerevisiae", FEMS Microbiology Letters, vol. 119, pp. 99-103, 1994. http://dx.doi.org/10.1111/j.1574-6968.1994.tb06874.x
- H. Kuriyama, and J. Slaughter, "Control of cell morphology of the yeast Saccharomyces cerevisiae by nutrient limitation in continuous culture", Letters in Applied Microbiology, vol. 20, pp. 37-40, 1995. http://dx.doi.org/10.1111/j.1472-765x.1995.tb00402.x
- A. Pitoniak, B. Birkaya, H.M. Dionne, N. Vadaie, and P.J. Cullen, "The Signaling Mucins Msb2 and Hkr1 Differentially Regulate the Filamentation Mitogen-activated Protein Kinase Pathway and Contribute to a Multimodal Response", Molecular Biology of the Cell, vol. 20, pp. 3101-3114, 2009. http://dx.doi.org/10.1091/mbc.E08-07-0760
- R. Martin, B. Wächtler, M. Schaller, D. Wilson, and B. Hube, "Host–pathogen interactions and virulence-associated genes during Candida albicans oral infections", International Journal of Medical Microbiology, vol. 301, pp. 417-422, 2011. http://dx.doi.org/10.1016/j.ijmm.2011.04.009
- S. Laxman, and B.P. Tu, "Multiple TORC1-Associated Proteins Regulate Nitrogen Starvation-Dependent Cellular Differentiation in Saccharomyces cerevisiae", PLoS ONE, vol. 6, pp. e26081, 2011. http://dx.doi.org/10.1371/journal.pone.0026081
- O. Ryan, R.S. Shapiro, C.F. Kurat, D. Mayhew, A. Baryshnikova, B. Chin, Z. Lin, M.J. Cox, F. Vizeacoumar, D. Cheung, S. Bahr, K. Tsui, F. Tebbji, A. Sellam, F. Istel, T. Schwarzmüller, T.B. Reynolds, K. Kuchler, D.K. Gifford, M. Whiteway, G. Giaever, C. Nislow, M. Costanzo, A. Gingras, R.D. Mitra, B. Andrews, G.R. Fink, L.E. Cowen, and C. Boone, "Global Gene Deletion Analysis Exploring Yeast Filamentous Growth", Science, vol. 337, pp. 1353-1356, 2012. http://dx.doi.org/10.1126/science.1224339
- J.D. Loeb, T.A. Kerentseva, T. Pan, M. Sepulveda-Becerra, and H. Liu, "Saccharomyces cerevisiae G1 cyclins are differentially involved in invasive and pseudohyphal growth independent of the filamentation mitogen-activated protein kinase pathway.", Genetics, 1999. http://www.ncbi.nlm.nih.gov/pubmed/10581264
- W. Lo, and A.M. Dranginis, "The Cell Surface Flocculin Flo11 Is Required for Pseudohyphae Formation and Invasion bySaccharomyces cerevisiae", Molecular Biology of the Cell, vol. 9, pp. 161-171, 1998. http://dx.doi.org/10.1091/mbc.9.1.161
-
K. Voordeckers, D. De Maeyer, E. van der Zande, M.D. Vinces, W. Meert, L. Cloots, O. Ryan, K. Marchal, and K.J. Verstrepen, "Identification of a complex genetic network underlying
S accharomyces cerevisiae colony morphology", Molecular Microbiology, vol. 86, pp. 225-239, 2012. http://dx.doi.org/10.1111/j.1365-2958.2012.08192.x - J.A. Granek, and P.M. Magwene, "Environmental and Genetic Determinants of Colony Morphology in Yeast", PLoS Genetics, vol. 6, pp. e1000823, 2010. http://dx.doi.org/10.1371/journal.pgen.1000823
- T.B. Reynolds, and G.R. Fink, "Bakers' Yeast, a Model for Fungal Biofilm Formation", Science, vol. 291, pp. 878-881, 2001. http://dx.doi.org/10.1126/science.291.5505.878
- G. Zara, S. Zara, C. Pinna, S. Marceddu, and M. Budroni, "FLO11 gene length and transcriptional level affect biofilm-forming ability of wild flor strains of Saccharomyces cerevisiae", Microbiology, vol. 155, pp. 3838-3846, 2009. http://dx.doi.org/10.1099/mic.0.028738-0
- L.M. Douglas, L. Li, Y. Yang, and A.M. Dranginis, "Expression and Characterization of the Flocculin Flo11/Muc1, a Saccharomyces cerevisiae Mannoprotein with Homotypic Properties of Adhesion", Eukaryotic Cell, vol. 6, pp. 2214-2221, 2007. http://dx.doi.org/10.1128/EC.00284-06
- P.J. Cullen, and G.F. Sprague, "The Regulation of Filamentous Growth in Yeast", Genetics, vol. 190, pp. 23-49, 2012. http://dx.doi.org/10.1534/genetics.111.127456
- S. Irniger, "The Ime2 protein kinase family in fungi: more duties than just meiosis", Molecular Microbiology, vol. 80, pp. 1-13, 2011. http://dx.doi.org/10.1111/j.1365-2958.2011.07575.x
- F. van Werven, G. Neuert, N. Hendrick, A. Lardenois, S. Buratowski, A. van Oudenaarden, M. Primig, and A. Amon, "Transcription of Two Long Noncoding RNAs Mediates Mating-Type Control of Gametogenesis in Budding Yeast", Cell, vol. 150, pp. 1170-1181, 2012. http://dx.doi.org/10.1016/j.cell.2012.06.049
- D. van Dyk, G. Hansson, I.S. Pretorius, and F.F. Bauer, "Cellular differentiation in response to nutrient availability: The repressor of meiosis, Rme1p, positively regulates invasive growth in Saccharomyces cerevisiae.", Genetics, 2003. http://www.ncbi.nlm.nih.gov/pubmed/14668363
- S. Rupp, "MAP kinase and cAMP filamentation signaling pathways converge on the unusually large promoter of the yeast FLO11 gene", The EMBO Journal, vol. 18, pp. 1257-1269, 1999. http://dx.doi.org/10.1093/emboj/18.5.1257
- J.C. Shah, and M.J. Clancy, "IME4, a gene that mediates MAT and nutritional control of meiosis in Saccharomyces cerevisiae.", Molecular and Cellular Biology, vol. 12, pp. 1078-1086, 1992. http://dx.doi.org/10.1128/mcb.12.3.1078
- C.F. Hongay, P.L. Grisafi, T. Galitski, and G.R. Fink, "Antisense Transcription Controls Cell Fate in Saccharomyces cerevisiae", Cell, vol. 127, pp. 735-745, 2006. http://dx.doi.org/10.1016/j.cell.2006.09.038
- B. Gelfand, J. Mead, A. Bruning, N. Apostolopoulos, V. Tadigotla, V. Nagaraj, A.M. Sengupta, and A.K. Vershon, "Regulated Antisense Transcription Controls Expression of Cell-Type-Specific Genes in Yeast", Molecular and Cellular Biology, vol. 31, pp. 1701-1709, 2011. http://dx.doi.org/10.1128/MCB.01071-10
- S.D. Agarwala, H.G. Blitzblau, A. Hochwagen, and G.R. Fink, "RNA Methylation by the MIS Complex Regulates a Cell Fate Decision in Yeast", PLoS Genetics, vol. 8, pp. e1002732, 2012. http://dx.doi.org/10.1371/journal.pgen.1002732
- S. Schwartz, S. Agarwala, M. Mumbach, M. Jovanovic, P. Mertins, A. Shishkin, Y. Tabach, T. Mikkelsen, R. Satija, G. Ruvkun, S. Carr, E. Lander, G. Fink, and A. Regev, "High-Resolution Mapping Reveals a Conserved, Widespread, Dynamic mRNA Methylation Program in Yeast Meiosis", Cell, vol. 155, pp. 1409-1421, 2013. http://dx.doi.org/10.1016/j.cell.2013.10.047
- Y. Yue, J. Liu, and C. He, "RNA N6-methyladenosine methylation in post-transcriptional gene expression regulation", Genes & Development, vol. 29, pp. 1343-1355, 2015. http://dx.doi.org/10.1101/gad.262766.115
- A.G. Uren, T. Beilharz, M.J. O’Connell, S.J. Bugg, R. van Driel, D.L. Vaux, and T. Lithgow, "Role for yeast inhibitor of apoptosis (IAP)-like proteins in cell division", Proceedings of the National Academy of Sciences, vol. 96, pp. 10170-10175, 1999. http://dx.doi.org/10.1073/pnas.96.18.10170
- S. Hurtado, K.S. Kim Guisbert, and E.J. Sontheimer, "SPO24 Is a Transcriptionally Dynamic, Small ORF-Encoding Locus Required for Efficient Sporulation in Saccharomyces cerevisiae", PLoS ONE, vol. 9, pp. e105058, 2014. http://dx.doi.org/10.1371/journal.pone.0105058
- B. Mai, and L.L. Breeden, "Identification of Target Genes of a Yeast Transcriptional Repressor", Differential Display Methods and Protocols, pp. 267-278, . http://dx.doi.org/10.1385/1-59259-968-0:267
- B. Mai, and L. Breeden, "CLN1 and Its Repression by Xbp1 Are Important for Efficient Sporulation in Budding Yeast", Molecular and Cellular Biology, vol. 20, pp. 478-487, 2000. http://dx.doi.org/10.1128/mcb.20.2.478-487.2000
- K. Purnapatre, S. Piccirillo, B.L. Schneider, and S.M. Honigberg, "The CLN3/SWI6/CLN2 pathway and SNF1 act sequentially to regulate meiotic initiation in Saccharomyces cerevisiae", Genes to Cells, vol. 7, pp. 675-691, 2002. http://dx.doi.org/10.1046/j.1365-2443.2002.00551.x
- N. Colomina, "G1 cyclins block the Ime1 pathway to make mitosis and meiosis incompatible in budding yeast", The EMBO Journal, vol. 18, pp. 320-329, 1999. http://dx.doi.org/10.1093/emboj/18.2.320
- A.D. Aragon, A.L. Rodriguez, O. Meirelles, S. Roy, G.S. Davidson, P.H. Tapia, C. Allen, R. Joe, D. Benn, and M. Werner-Washburne, "Characterization of Differentiated Quiescent and Nonquiescent Cells in Yeast Stationary-Phase Cultures", Molecular Biology of the Cell, vol. 19, pp. 1271-1280, 2008. http://dx.doi.org/10.1091/mbc.E07-07-0666
- L. Shi, B.M. Sutter, X. Ye, and B.P. Tu, "Trehalose Is a Key Determinant of the Quiescent Metabolic State That Fuels Cell Cycle Progression upon Return to Growth", Molecular Biology of the Cell, vol. 21, pp. 1982-1990, 2010. http://dx.doi.org/10.1091/mbc.E10-01-0056
- H. Tapia, and K.A. Morano, "Hsp90 Nuclear Accumulation in Quiescence Is Linked to Chaperone Function and Spore Development in Yeast", Molecular Biology of the Cell, vol. 21, pp. 63-72, 2010. http://dx.doi.org/10.1091/mbc.E09-05-0376
- S. Kurtz, J. Rossi, L. Petko, and S. Lindquist, "An Ancient Developmental Induction: Heat-Shock Proteins Induced in Sporulation and Oogenesis", Science, vol. 231, pp. 1154-1157, 1986. http://dx.doi.org/10.1126/science.3511530
- S.M. Honigberg, and K. Purnapatre, "Signal pathway integration in the switch from the mitotic cell cycle to meiosis in yeast", Journal of Cell Science, vol. 116, pp. 2137-2147, 2003. http://dx.doi.org/10.1242/jcs.00460
- F.J. van Werven, and A. Amon, "Regulation of entry into gametogenesis", Philosophical Transactions of the Royal Society B: Biological Sciences, vol. 366, pp. 3521-3531, 2011. http://dx.doi.org/10.1098/rstb.2011.0081
- M. Werner-Washburne, S. Roy, and G.S. Davidson, "Aging and the Survival of Quiescent and Non-quiescent Cells in Yeast Stationary-Phase Cultures", Aging Research in Yeast, pp. 123-143, 2011. http://dx.doi.org/10.1007/978-94-007-2561-4_6
- S.R. Ceccato-Antonini, and P.E. Sudbery, "Filamentous growth in Saccharomyces cerevisiae", Brazilian Journal of Microbiology, vol. 35, pp. 173-181, 2004. http://dx.doi.org/10.1590/s1517-83822004000200001
- A. Jambhekar, and A. Amon, "Control of Meiosis by Respiration", Current Biology, vol. 18, pp. 969-975, 2008. http://dx.doi.org/10.1016/j.cub.2008.05.047
- R.H. Lee, and S.M. Honigberg, "Nutritional Regulation of Late Meiotic Events in Saccharomyces cerevisiae through a Pathway Distinct from Initiation†", Molecular and Cellular Biology, vol. 16, pp. 3222-3232, 1996. http://dx.doi.org/10.1128/mcb.16.6.3222
- M.K. Conway, D. Grunwald, and W. Heideman, "Glucose, Nitrogen, and Phosphate Repletion in Saccharomyces cerevisiae: Common Transcriptional Responses to Different Nutrient Signals", G3 Genes|Genomes|Genetics, vol. 2, pp. 1003-1017, 2012. http://dx.doi.org/10.1534/g3.112.002808
- L. Galdieri, S. Mehrotra, S. Yu, and A. Vancura, "Transcriptional Regulation in Yeast during Diauxic Shift and Stationary Phase", OMICS: A Journal of Integrative Biology, vol. 14, pp. 629-638, 2010. http://dx.doi.org/10.1089/omi.2010.0069
- M.M. Klosinska, C.A. Crutchfield, P.H. Bradley, J.D. Rabinowitz, and J.R. Broach, "Yeast cells can access distinct quiescent states", Genes & Development, vol. 25, pp. 336-349, 2011. http://dx.doi.org/10.1101/gad.2011311
- I. Orlandi, R. Ronzulli, N. Casatta, and M. Vai, "Ethanol and Acetate Acting as Carbon/Energy Sources Negatively Affect Yeast Chronological Aging", Oxidative Medicine and Cellular Longevity, vol. 2013, pp. 1-10, 2013. http://dx.doi.org/10.1155/2013/802870
- C.R. Burtner, C.J. Murakami, B.K. Kennedy, and M. Kaeberlein, "A molecular mechanism of chronological aging in yeast", Cell Cycle, vol. 8, pp. 1256-1270, 2009. http://dx.doi.org/10.4161/cc.8.8.8287
- S.P. Palecek, A.S. Parikh, J.H. Huh, and S.J. Kron, "Depression of Saccharomyces cerevisiae invasive growth on non‐glucose carbon sources requires the Snf1 kinase", Molecular Microbiology, vol. 45, pp. 453-469, 2002. http://dx.doi.org/10.1046/j.1365-2958.2002.03024.x
- P.J. Cullen, and G.F. Sprague, "The Roles of Bud-Site-Selection Proteins during Haploid Invasive Growth in Yeast", Molecular Biology of the Cell, vol. 13, pp. 2990-3004, 2002. http://dx.doi.org/10.1091/mbc.E02-03-0151
- P.J. Cullen, "Evaluating Yeast Filamentous Growth at the Single-Cell Level", Cold Spring Harbor Protocols, vol. 2015, pp. pdb.prot085084, 2015. http://dx.doi.org/10.1101/pdb.prot085084
- P. Godard, A. Urrestarazu, S. Vissers, K. Kontos, G. Bontempi, J. van Helden, and B. André, "Effect of 21 Different Nitrogen Sources on Global Gene Expression in the YeastSaccharomyces cerevisiae", Molecular and Cellular Biology, vol. 27, pp. 3065-3086, 2007. http://dx.doi.org/10.1128/MCB.01084-06
- A. Day, J. Markwardt, R. Delaguila, J. Zhang, K. Purnapatre, S.M. Honigberg, and B.L. Schneider, "Cell Size and Cln-Cdc28 Complexes Mediate Entry into Meiosis by Modulating Cell Growth", Cell Cycle, vol. 3, pp. 1433-1439, 2004. http://dx.doi.org/10.4161/cc.3.11.1205
- N. Colomina, Y. Liu, M. Aldea, and E. Garí, "TOR Regulates the Subcellular Localization of Ime1, a Transcriptional Activator of Meiotic Development in Budding Yeast", Molecular and Cellular Biology, vol. 23, pp. 7415-7424, 2003. http://dx.doi.org/10.1128/mcb.23.20.7415-7424.2003
- J. Santos, F. Leitão-Correia, M.J. Sousa, and C. Leão, "Ammonium is a key determinant on the dietary restriction of yeast chronological aging in culture medium", Oncotarget, vol. 6, pp. 6511-6523, 2014. http://dx.doi.org/10.18632/oncotarget.2989
- P.K. Vinod, N. Sengupta, P.J. Bhat, and K.V. Venkatesh, "Integration of Global Signaling Pathways, cAMP-PKA, MAPK and TOR in the Regulation of FLO11", PLoS ONE, vol. 3, pp. e1663, 2008. http://dx.doi.org/10.1371/journal.pone.0001663
- M. Boeckstaens, B. André, and A.M. Marini, "The yeast ammonium transport protein Mep2 and its positive regulator, the Npr1 kinase, play an important role in normal and pseudohyphal growth on various nitrogen media through retrieval of excreted ammonium", Molecular Microbiology, vol. 64, pp. 534-546, 2007. http://dx.doi.org/10.1111/j.1365-2958.2007.05681.x
- L.E. Murray, N. Rowley, I.W. Dawes, G.C. Johnston, and R.A. Singer, "A yeast glutamine tRNA signals nitrogen status for regulation of dimorphic growth and sporulation", Proceedings of the National Academy of Sciences, vol. 95, pp. 8619-8624, 1998. http://dx.doi.org/10.1073/pnas.95.15.8619
- M.C. Lorenz, "The MEP2 ammonium permease regulates pseudohyphal differentiation in Saccharomyces cerevisiae", The EMBO Journal, vol. 17, pp. 1236-1247, 1998. http://dx.doi.org/10.1093/emboj/17.5.1236
- R. Orij, J. Postmus, A. Ter Beek, S. Brul, and G.J. Smits, "In vivo measurement of cytosolic and mitochondrial pH using a pH-sensitive GFP derivative in Saccharomyces cerevisiae reveals a relation between intracellular pH and growth", Microbiology, vol. 155, pp. 268-278, 2009. http://dx.doi.org/10.1099/mic.0.022038-0
- C. Murakami, J.R. Delaney, A. Chou, D. Carr, J. Schleit, G.L. Sutphin, E.H. An, A.S. Castanza, M. Fletcher, S. Goswami, S. Higgins, M. Holmberg, J. Hui, M. Jelic, K. Jeong, J.R. Kim, S. Klum, E. Liao, M.S. Lin, W. Lo, H. Miller, R. Moller, Z.J. Peng, T. Pollard, P. Pradeep, D. Pruett, D. Rai, V. Ros, A. Schuster, M. Singh, B.L. Spector, H. Vander Wende, A.M. Wang, B.M. Wasko, B. Olsen, and M. Kaeberlein, "pH neutralization protects against reduction in replicative lifespan following chronological aging in yeast", Cell Cycle, vol. 11, pp. 3087-3096, 2012. http://dx.doi.org/10.4161/cc.21465
- M. Hayashi, K. Ohkuni, and I. Yamashita, "Control of division arrest and entry into meiosis by extracellular alkalisation in Saccharomyces cerevisiae.", Yeast (Chichester, England), 1998. http://www.ncbi.nlm.nih.gov/pubmed/9717236
- M.G. Mirisola, and V.D. Longo, "Acetic acid and acidification accelerate chronological and replicative aging in yeast", Cell Cycle, vol. 11, pp. 3532-3533, 2012. http://dx.doi.org/10.4161/cc.22042
- S. Piccirillo, M.G. White, J.C. Murphy, D.J. Law, and S.M. Honigberg, "The Rim101p/PacC Pathway and Alkaline pH Regulate Pattern Formation in Yeast Colonies", Genetics, vol. 184, pp. 707-716, 2010. http://dx.doi.org/10.1534/genetics.109.113480
- K. Ohkuni, M. Hayashi, and I. Yamashita, "Bicarbonate-mediated social communication stimulates meiosis and sporulation of Saccharomyces cerevisiae.", Yeast (Chichester, England), 1998. http://www.ncbi.nlm.nih.gov/pubmed/9639309
- E. Soares, "Flocculation in Saccharomyces cerevisiae: a review", Journal of Applied Microbiology, vol. 110, pp. 1-18, 2010. http://dx.doi.org/10.1111/j.1365-2672.2010.04897.x
- M. Stratford, "Induction of flocculation in brewing yeasts by change in pH value", FEMS Microbiology Letters, vol. 136, pp. 13-18, 1996. http://dx.doi.org/10.1111/j.1574-6968.1996.tb08018.x
- W. Li, and A.P. Mitchell, "Proteolytic activation of Rim1p, a positive regulator of yeast sporulation and invasive growth.", Genetics, 1997. http://www.ncbi.nlm.nih.gov/pubmed/9017390
- M.S. Esposito, and R.E. Esposito, "The genetic control of sporulation in Saccharomyces. I. The isolation of temperature-sensitive sporulation-deficient mutants.", Genetics, 1969. http://www.ncbi.nlm.nih.gov/pubmed/5802566
- J.H. McCusker, K.V. Clemons, D.A. Stevens, and R.W. Davis, "Saccharomyces cerevisiae virulence phenotype as determined with CD-1 mice is associated with the ability to grow at 42 degrees C and form pseudohyphae.", Infection and immunity, 1994. http://www.ncbi.nlm.nih.gov/pubmed/7960125
- J.H. McCusker, K.V. Clemons, D.A. Stevens, and R.W. Davis, "Genetic characterization of pathogenic Saccharomyces cerevisiae isolates.", Genetics, 1994. http://www.ncbi.nlm.nih.gov/pubmed/8013903
- I. Nussbaum, E. Weindling, R. Jubran, A. Cohen, and S. Bar-Nun, "Deteriorated Stress Response in Stationary-Phase Yeast: Sir2 and Yap1 Are Essential for Hsf1 Activation by Heat Shock and Oxidative Stress, Respectively", PLoS ONE, vol. 9, pp. e111505, 2014. http://dx.doi.org/10.1371/journal.pone.0111505
- N. Kourtis, and N. Tavernarakis, "Cellular stress response pathways and ageing: intricate molecular relationships", The EMBO Journal, vol. 30, pp. 2520-2531, 2011. http://dx.doi.org/10.1038/emboj.2011.162
- K.J. Verstrepen, and G.R. Fink, "Genetic and Epigenetic Mechanisms Underlying Cell-Surface Variability in Protozoa and Fungi", Annual Review of Genetics, vol. 43, pp. 1-24, 2009. http://dx.doi.org/10.1146/annurev-genet-102108-134156
- P.J. Cullen, "Signaling Mucins: The New Kids on the MARK Block", Critical Reviews™ in Eukaryotic Gene Expression, vol. 17, pp. 241-257, 2007. http://dx.doi.org/10.1615/critreveukargeneexpr.v17.i3.50
- M. Conrad, J. Schothorst, H.N. Kankipati, G. Van Zeebroeck, M. Rubio-Texeira, and J.M. Thevelein, "Nutrient sensing and signaling in the yeastSaccharomyces cerevisiae", FEMS Microbiology Reviews, vol. 38, pp. 254-299, 2014. http://dx.doi.org/10.1111/1574-6976.12065
- J.R. Broach, "Nutritional Control of Growth and Development in Yeast", Genetics, vol. 192, pp. 73-105, 2012. http://dx.doi.org/10.1534/genetics.111.135731
- K. Hedbacker, "SNF1/AMPK pathways in yeast", Frontiers in Bioscience, vol. 13, pp. 2408, 2008. http://dx.doi.org/10.2741/2854
- M. Orlova, H. Ozcetin, L. Barrett, and S. Kuchin, "Roles of the Snf1-Activating Kinases during Nitrogen Limitation and Pseudohyphal Differentiation in Saccharomyces cerevisiae", Eukaryotic Cell, vol. 9, pp. 208-214, 2010. http://dx.doi.org/10.1128/EC.00216-09
- J.E. Hughes Hallett, X. Luo, and A.P. Capaldi, "State Transitions in the TORC1 Signaling Pathway and Information Processing in Saccharomyces cerevisiae", Genetics, vol. 198, pp. 773-786, 2014. http://dx.doi.org/10.1534/genetics.114.168369
- J. Thevelein, B. Bonini, D. Castermans, S. Haesendonckx, J. Kriel, W. Louwet, P. Thayumanavan, Y. Popova, M. Rubio-Texeira, W. Schepers, P. Vandormael, G. Zeebroeck, P. Verhaert, M. Versele, and K. Voordeckers, "Novel mechanisms in nutrient activation of the yeast Protein Kinase A pathway", Acta Microbiologica et Immunologica Hungarica, vol. 55, pp. 75-89, 2008. http://dx.doi.org/10.1556/AMicr.55.2008.2.1
- M. Jungbluth, H. Mösch, and C. Taxis, "Acetate Regulation of Spore Formation Is under the Control of the Ras/Cyclic AMP/Protein Kinase A Pathway and Carbon Dioxide in Saccharomyces cerevisiae", Eukaryotic Cell, vol. 11, pp. 1021-1032, 2012. http://dx.doi.org/10.1128/EC.05240-11
- R. Dechant, S. Saad, A. Ibáñez, and M. Peter, "Cytosolic pH Regulates Cell Growth through Distinct GTPases, Arf1 and Gtr1, to Promote Ras/PKA and TORC1 Activity", Molecular Cell, vol. 55, pp. 409-421, 2014. http://dx.doi.org/10.1016/j.molcel.2014.06.002
- R. Dechant, M. Binda, S.S. Lee, S. Pelet, J. Winderickx, and M. Peter, "Cytosolic pH is a second messenger for glucose and regulates the PKA pathway through V-ATPase", The EMBO Journal, vol. 29, pp. 2515-2526, 2010. http://dx.doi.org/10.1038/emboj.2010.138
- M. Mizunuma, R. Tsubakiyama, T. Ogawa, A. Shitamukai, Y. Kobayashi, T. Inai, K. Kume, and D. Hirata, "Ras/cAMP-dependent Protein Kinase (PKA) Regulates Multiple Aspects of Cellular Events by Phosphorylating the Whi3 Cell Cycle Regulator in Budding Yeast", Journal of Biological Chemistry, vol. 288, pp. 10558-10566, 2013. http://dx.doi.org/10.1074/jbc.M112.402214
- C.R. Burtner, C.J. Murakami, B. Olsen, B.K. Kennedy, and M. Kaeberlein, "A genomic analysis of chronological longevity factors in budding yeast", Cell Cycle, vol. 10, pp. 1385-1396, 2011. http://dx.doi.org/10.4161/cc.10.9.15464
- H.U. Mösch, and G.R. Fink, "Dissection of filamentous growth by transposon mutagenesis in Saccharomyces cerevisiae.", Genetics, 1997. http://www.ncbi.nlm.nih.gov/pubmed/9055077
- A. Reinders, N. Bürckert, T. Boller, A. Wiemken, and C. De Virgilio, "Saccharomyces cerevisiae cAMP-dependent protein kinase controls entry into stationary phase through the Rim15p protein kinase", Genes & Development, vol. 12, pp. 2943-2955, 1998. http://dx.doi.org/10.1101/gad.12.18.2943
- D. Watanabe, Y. Zhou, A. Hirata, Y. Sugimoto, K. Takagi, T. Akao, Y. Ohya, H. Takagi, and H. Shimoi, "Inhibitory Role of Greatwall-Like Protein Kinase Rim15p in Alcoholic Fermentation via Upregulating the UDP-Glucose Synthesis Pathway in Saccharomyces cerevisiae", Applied and Environmental Microbiology, vol. 82, pp. 340-351, 2016. http://dx.doi.org/10.1128/AEM.02977-15
- S. Bontron, M. Jaquenoud, S. Vaga, N. Talarek, B. Bodenmiller, R. Aebersold, and C. De Virgilio, "Yeast Endosulfines Control Entry into Quiescence and Chronological Life Span by Inhibiting Protein Phosphatase 2A", Cell Reports, vol. 3, pp. 16-22, 2013. http://dx.doi.org/10.1016/j.celrep.2012.11.025
- X. Luo, N. Talarek, and C. De Virgilio, "Initiation of the yeast G0program requires Igo1 and Igo2, which antagonize activation of decapping of specific nutrient-regulated mRNAs", RNA Biology, vol. 8, pp. 14-17, 2011. http://dx.doi.org/10.4161/rna.8.1.13483
- N. Talarek, E. Cameroni, M. Jaquenoud, X. Luo, S. Bontron, S. Lippman, G. Devgan, M. Snyder, J.R. Broach, and C. De Virgilio, "Initiation of the TORC1-Regulated G0 Program Requires Igo1/2, which License Specific mRNAs to Evade Degradation via the 5′-3′ mRNA Decay Pathway", Molecular Cell, vol. 38, pp. 345-355, 2010. http://dx.doi.org/10.1016/j.molcel.2010.02.039
- P. Lee, M.S. Kim, S. Paik, S. Choi, B. Cho, and J. Hahn, "Rim15‐dependent activation of Hsf1 and Msn2/4 transcription factors by direct phosphorylation in Saccharomyces cerevisiae", FEBS Letters, vol. 587, pp. 3648-3655, 2013. http://dx.doi.org/10.1016/j.febslet.2013.10.004
- M. Wei, P. Fabrizio, J. Hu, H. Ge, C. Cheng, L. Li, and V.D. Longo, "Life Span Extension by Calorie Restriction Depends on Rim15 and Transcription Factors Downstream of Ras/PKA, Tor, and Sch9", PLoS Genetics, vol. 4, pp. e13, 2008. http://dx.doi.org/10.1371/journal.pgen.0040013
- S. Vidan, and A.P. Mitchell, "Stimulation of Yeast Meiotic Gene Expression by the Glucose-Repressible Protein Kinase Rim15p", Molecular and Cellular Biology, vol. 17, pp. 2688-2697, 1997. http://dx.doi.org/10.1128/mcb.17.5.2688
- Z. Quan, L. Cao, Y. Tang, Y. Yan, S.G. Oliver, and N. Zhang, "The Yeast GSK-3 Homologue Mck1 Is a Key Controller of Quiescence Entry and Chronological Lifespan", PLOS Genetics, vol. 11, pp. e1005282, 2015. http://dx.doi.org/10.1371/journal.pgen.1005282
- E. Swinnen, V. Wanke, J. Roosen, B. Smets, F. Dubouloz, I. Pedruzzi, E. Cameroni, C. De Virgilio, and J. Winderickx, "Rim15 and the crossroads of nutrient signalling pathways in Saccharomyces cerevisiae", Cell Division, vol. 1, 2006. http://dx.doi.org/10.1186/1747-1028-1-3
- S. Sarkar, J.Z. Dalgaard, J.B.A. Millar, and P. Arumugam, "The Rim15-Endosulfine-PP2ACdc55 Signalling Module Regulates Entry into Gametogenesis and Quiescence via Distinct Mechanisms in Budding Yeast", PLoS Genetics, vol. 10, pp. e1004456, 2014. http://dx.doi.org/10.1371/journal.pgen.1004456
- M.P. Ward, C.J. Gimeno, G.R. Fink, and S. Garrett, "SOK2 May Regulate Cyclic AMP-Dependent Protein Kinase-Stimulated Growth and Pseudohyphal", Molecular and Cellular Biology, vol. 15, pp. 6854-6863, 1995. http://dx.doi.org/10.1128/mcb.15.12.6854
- M.P. Ward, and S. Garrett, "Suppression of a yeast cyclic AMP-dependent protein kinase defect by overexpression of SOK1, a yeast gene exhibiting sequence similarity to a developmentally regulated mouse gene.", Molecular and Cellular Biology, vol. 14, pp. 5619-5627, 1994. http://dx.doi.org/10.1128/mcb.14.9.5619
- G. Shenhar, and Y. Kassir, "A Positive Regulator of Mitosis, Sok2, Functions as a Negative Regulator of Meiosis in Saccharomyces cerevisiae", Molecular and Cellular Biology, vol. 21, pp. 1603-1612, 2001. http://dx.doi.org/10.1128/MCB.21.5.1603-1612.2001
- X. Pan, and J. Heitman, "Sok2 Regulates Yeast Pseudohyphal Differentiation via a Transcription Factor Cascade That Regulates Cell-Cell Adhesion", Molecular and Cellular Biology, vol. 20, pp. 8364-8372, 2000. http://dx.doi.org/10.1128/mcb.20.22.8364-8372.2000
- A.R. Borneman, J.A. Leigh-Bell, H. Yu, P. Bertone, M. Gerstein, and M. Snyder, "Target hub proteins serve as master regulators of development in yeast", Genes & Development, vol. 20, pp. 435-448, 2006. http://dx.doi.org/10.1101/gad.1389306
- C.M. McDonald, M. Wagner, M.J. Dunham, M.E. Shin, N.T. Ahmed, and E. Winter, "The Ras/cAMP Pathway and the CDK-Like Kinase Ime2 Regulate the MAPK Smk1 and Spore Morphogenesis in Saccharomyces cerevisiae", Genetics, vol. 181, pp. 511-523, 2009. http://dx.doi.org/10.1534/genetics.108.098434
- P. Lee, S. Paik, C. Shin, W. Huh, and J. Hahn, "Regulation of yeast Yak1 kinase by PKA and autophosphorylation-dependent 14-3-3 binding", Molecular Microbiology, vol. 79, pp. 633-646, 2010. http://dx.doi.org/10.1111/j.1365-2958.2010.07471.x
- P. Lee, B. Cho, H. Joo, and J. Hahn, "Yeast Yak1 kinase, a bridge between PKA and stress‐responsive transcription factors, Hsf1 and Msn2/Msn4", Molecular Microbiology, vol. 70, pp. 882-895, 2008. http://dx.doi.org/10.1111/j.1365-2958.2008.06450.x
- F. Cardona, M. Olmo, and A. Aranda, "Phylogenetic origin and transcriptional regulation at the post-diauxic phase of SPI1, in Saccharomyces cerevisiae", Cellular and Molecular Biology Letters, vol. 17, 2012. http://dx.doi.org/10.2478/s11658-012-0017-4
- M. Werner-Washburne, D. Brown, and E. Braun, "Bcy1, the regulatory subunit of cAMP-dependent protein kinase in yeast, is differentially modified in response to the physiological status of the cell.", The Journal of biological chemistry, 1991. http://www.ncbi.nlm.nih.gov/pubmed/1655793
- M. Malcher, S. Schladebeck, and H. Mösch, "The Yak1 Protein Kinase Lies at the Center of a Regulatory Cascade Affecting Adhesive Growth and Stress Resistance inSaccharomyces cerevisiae", Genetics, vol. 187, pp. 717-730, 2011. http://dx.doi.org/10.1534/genetics.110.125708
- M.G. Mirisola, G. Taormina, P. Fabrizio, M. Wei, J. Hu, and V.D. Longo, "Serine- and Threonine/Valine-Dependent Activation of PDK and Tor Orthologs Converge on Sch9 to Promote Aging", PLoS Genetics, vol. 10, pp. e1004113, 2014. http://dx.doi.org/10.1371/journal.pgen.1004113
- E. Swinnen, R. Ghillebert, T. Wilms, and J. Winderickx, "Molecular mechanisms linking the evolutionary conserved TORC1-Sch9 nutrient signalling branch to lifespan regulation inSaccharomyces cerevisiae", FEMS Yeast Research, vol. 14, pp. 17-32, 2013. http://dx.doi.org/10.1111/1567-1364.12097
- H. Weidberg, F. Moretto, G. Spedale, A. Amon, and F.J. van Werven, "Nutrient Control of Yeast Gametogenesis Is Mediated by TORC1, PKA and Energy Availability", PLOS Genetics, vol. 12, pp. e1006075, 2016. http://dx.doi.org/10.1371/journal.pgen.1006075
- Y. Pan, E. Schroeder, A. Ocampo, A. Barrientos, and G. Shadel, "Regulation of Yeast Chronological Life Span by TORC1 via Adaptive Mitochondrial ROS Signaling", Cell Metabolism, vol. 13, pp. 668-678, 2011. http://dx.doi.org/10.1016/j.cmet.2011.03.018
- Y. Pan, and G.S. Shadel, "Extension of chronological life span by reduced TOR signaling requires down-regulation of Sch9p and involves increased mitochondrial OXPHOS complex density", Aging, vol. 1, pp. 131-145, 2009. http://dx.doi.org/10.18632/aging.100016
- N.S. Cutler, X. Pan, J. Heitman, and M.E. Cardenas, "The TOR Signal Transduction Cascade Controls Cellular Differentiation in Response to Nutrients", Molecular Biology of the Cell, vol. 12, pp. 4103-4113, 2001. http://dx.doi.org/10.1091/mbc.12.12.4103
- S. Brückner, S. Kern, R. Birke, I. Saugar, H.D. Ulrich, and H. Mösch, "The TEA Transcription Factor Tec1 Links TOR and MAPK Pathways to Coordinate Yeast Development", Genetics, vol. 189, pp. 479-494, 2011. http://dx.doi.org/10.1534/genetics.111.133629
- E.M. Humston, K.M. Dombek, B.P. Tu, E.T. Young, and R.E. Synovec, "Toward a global analysis of metabolites in regulatory mutants of yeast", Analytical and Bioanalytical Chemistry, vol. 401, pp. 2387-2402, 2011. http://dx.doi.org/10.1007/s00216-011-4800-2
- R. Usaite, M.C. Jewett, A.P. Oliveira, J.R. Yates, L. Olsson, and J. Nielsen, "Reconstruction of the yeast Snf1 kinase regulatory network reveals its role as a global energy regulator", Molecular Systems Biology, vol. 5, 2009. http://dx.doi.org/10.1038/msb.2009.67
- Z. Wang, W.A. Wilson, M.A. Fujino, and P.J. Roach, "Antagonistic Controls of Autophagy and Glycogen Accumulation by Snf1p, the Yeast Homolog of AMP-Activated Protein Kinase, and the Cyclin-Dependent Kinase Pho85p", Molecular and Cellular Biology, vol. 21, pp. 5742-5752, 2001. http://dx.doi.org/10.1128/mcb.21.17.5742-5752.2001
- B. Enjalbert, J.L. Parrou, M.A. Teste, and J. François, "Combinatorial control by the protein kinases PKA, PHO85 and SNF1 of transcriptional induction of the Saccharomyces cerevisiae GSY2 gene at the diauxic shift", Molecular Genetics and Genomics, vol. 271, pp. 697-708, 2004. http://dx.doi.org/10.1007/s00438-004-1014-8
- S. Van de Velde, and J.M. Thevelein, "Cyclic AMP-Protein Kinase A and Snf1 Signaling Mechanisms Underlie the Superior Potency of Sucrose for Induction of Filamentation in Saccharomyces cerevisiae", Eukaryotic Cell, vol. 7, pp. 286-293, 2008. http://dx.doi.org/10.1128/EC.00276-07
- S. Kuchin, V.K. Vyas, and M. Carlson, "Snf1 Protein Kinase and the Repressors Nrg1 and Nrg2 Regulate FLO11, Haploid Invasive Growth, and Diploid Pseudohyphal Differentiation", Molecular and Cellular Biology, vol. 22, pp. 3994-4000, 2002. http://dx.doi.org/10.1128/mcb.22.12.3994-4000.2002
- D.R. Lorenz, C.R. Cantor, and J.J. Collins, "A network biology approach to aging in yeast", Proceedings of the National Academy of Sciences, vol. 106, pp. 1145-1150, 2009. http://dx.doi.org/10.1073/pnas.0812551106
- K. Ashrafi, S.S. Lin, J.K. Manchester, and J.I. Gordon, "Sip2p and its partner snf1p kinase affect aging in S. cerevisiae.", Genes & development, 2000. http://www.ncbi.nlm.nih.gov/pubmed/10921902
- T. Maeda, "The signaling mechanism of ambient pH sensing and adaptation in yeast and fungi", The FEBS Journal, vol. 279, pp. 1407-1413, 2012. http://dx.doi.org/10.1111/j.1742-4658.2012.08548.x
- K.S. Andersen, R. Bojsen, L.G.R. Sørensen, M.W. Nielsen, M. Lisby, A. Folkesson, and B. Regenberg, "Genetic Basis for Saccharomyces cerevisiae Biofilm in Liquid Medium", G3 Genes|Genomes|Genetics, vol. 4, pp. 1671-1680, 2014. http://dx.doi.org/10.1534/g3.114.010892
- C.A. Chavel, H.M. Dionne, B. Birkaya, J. Joshi, and P.J. Cullen, "Multiple Signals Converge on a Differentiation MAPK Pathway", PLoS Genetics, vol. 6, pp. e1000883, 2010. http://dx.doi.org/10.1371/journal.pgen.1000883
- T.M. Lamb, and A.P. Mitchell, "The Transcription Factor Rim101p Governs Ion Tolerance and Cell Differentiation by Direct Repression of the Regulatory Genes NRG1 and SMP1 in Saccharomyces cerevisiae", Molecular and Cellular Biology, vol. 23, pp. 677-686, 2003. http://dx.doi.org/10.1128/mcb.23.2.677-686.2003
- T. Read, P.A. Richmond, and R.D. Dowell, "A trans-acting Variant within the Transcription Factor RIM101 Interacts with Genetic Background to Determine its Regulatory Capacity", PLOS Genetics, vol. 12, pp. e1005746, 2016. http://dx.doi.org/10.1371/journal.pgen.1005746
- M. Gray, S. Piccirillo, K. Purnapatre, B.L. Schneider, and S.M. Honigberg, "Glucose induction pathway regulates meiosis inSaccharomyces cerevisiaein part by controlling turnover of Ime2p meiotic kinase", FEMS Yeast Research, vol. 8, pp. 676-684, 2008. http://dx.doi.org/10.1111/j.1567-1364.2008.00406.x
- K. Purnapatre, M. Gray, S. Piccirillo, and S.M. Honigberg, "Glucose Inhibits Meiotic DNA Replication through SCFGrr1p-Dependent Destruction of Ime2p Kinase", Molecular and Cellular Biology, vol. 25, pp. 440-450, 2005. http://dx.doi.org/10.1128/MCB.25.1.440-450.2005
- C.A. Shively, M.J. Eckwahl, C.J. Dobry, D. Mellacheruvu, A. Nesvizhskii, and A. Kumar, "Genetic Networks Inducing Invasive Growth in Saccharomyces cerevisiae Identified Through Systematic Genome-Wide Overexpression", Genetics, vol. 193, pp. 1297-1310, 2013. http://dx.doi.org/10.1534/genetics.112.147876
- K. Furukawa, T. Furukawa, and S. Hohmann, "Efficient Construction of Homozygous Diploid Strains Identifies Genes Required for the Hyper-Filamentous Phenotype in Saccharomyces cerevisiae", PLoS ONE, vol. 6, pp. e26584, 2011. http://dx.doi.org/10.1371/journal.pone.0026584
- S.A. Krause, and J.V. Gray, "The Protein Kinase C Pathway Is Required for Viability in Quiescence in Saccharomyces cerevisiae", Current Biology, vol. 12, pp. 588-593, 2002. http://dx.doi.org/10.1016/s0960-9822(02)00760-1
-
V. Sundaram, M.I. Petkova, N. Pujol‐Carrion, J. Boada, and M.A. de la Torre‐Ruiz, "
T or1,S ch9 andPKA downregulation in quiescence rely onM tl1 to preserve mitochondrial integrity and cell survival", Molecular Microbiology, vol. 97, pp. 93-109, 2015. http://dx.doi.org/10.1111/mmi.13013 - G.T. Vancetto, and S.R. Ceccato-Antonini, "MPK1 gene is required for filamentous growth induced by isoamyl alcohol in Saccharomyces cerevisiae strains from the alcoholic fermentation", Applied Microbiology and Biotechnology, vol. 75, pp. 111-115, 2007. http://dx.doi.org/10.1007/s00253-006-0795-6
- I.M. de, P. Marechal, and P. Gervais, "Passive Response ofSaccharomyces cerevisiaeto Osmotic Shifts: Cell Volume Variations Depending on the Physiological State", Biochemical and Biophysical Research Communications, vol. 227, pp. 519-523, 1996. http://dx.doi.org/10.1006/bbrc.1996.1539
- R. de Llanos, C. Hernández‐Haro, E. Barrio, A. Querol, M.T. Fernández‐Espinar, and M. Molina, "Differences in activation of MAP kinases and variability in the polyglutamine tract of Slt2 in clinical and non‐clinical isolates of Saccharomyces cerevisiae", Yeast, vol. 27, pp. 549-561, 2010. http://dx.doi.org/10.1002/yea.1799
- M. Schröder, R. Clark, C.Y. Liu, and R.J. Kaufman, "The unfolded protein response represses differentiation through the RPD3-SIN3 histone deacetylase", The EMBO Journal, vol. 23, pp. 2281-2292, 2004. http://dx.doi.org/10.1038/sj.emboj.7600233
- M. Orlova, E. Kanter, D. Krakovich, and S. Kuchin, "Nitrogen Availability and TOR Regulate the Snf1 Protein Kinase in Saccharomyces cerevisiae", Eukaryotic Cell, vol. 5, pp. 1831-1837, 2006. http://dx.doi.org/10.1128/EC.00110-06
- M. Nishizawa, M. Tanigawa, M. Hayashi, T. Maeda, Y. Yazaki, Y. Saeki, and A. Toh-e, "Pho85 Kinase, a Cyclin-Dependent Kinase, Regulates Nuclear Accumulation of the Rim101 Transcription Factor in the Stress Response of Saccharomyces cerevisiae", Eukaryotic Cell, vol. 9, pp. 943-951, 2010. http://dx.doi.org/10.1128/EC.00247-09
- M.J. Law, and K. Ciccaglione, "Fine-Tuning of Histone H3 Lys4 Methylation During Pseudohyphal Differentiation by the CDK Submodule of RNA Polymerase II", Genetics, vol. 199, pp. 435-453, 2014. http://dx.doi.org/10.1534/genetics.114.172841
- A. Halme, S. Bumgarner, C. Styles, and G.R. Fink, "Genetic and Epigenetic Regulation of the FLO Gene Family Generates Cell-Surface Variation in Yeast", Cell, vol. 116, pp. 405-415, 2004. http://dx.doi.org/10.1016/s0092-8674(04)00118-7
- L. Wang, F. Montalvo-Munoz, Y. Tsai, C. Liang, C. Chang, and W. Lo, "The Histone Acetyltransferase Gcn5 Regulates ncRNA-ICR1andFLO11Expression during Pseudohyphal Development inSaccharomyces cerevisiae", BioMed Research International, vol. 2015, pp. 1-13, 2015. http://dx.doi.org/10.1155/2015/284692
- S. Bumgarner, G. Neuert, B. Voight, A. Symbor-Nagrabska, P. Grisafi, A. van Oudenaarden, and G. Fink, "Single-Cell Analysis Reveals that Noncoding RNAs Contribute to Clonal Heterogeneity by Modulating Transcription Factor Recruitment", Molecular Cell, vol. 45, pp. 470-482, 2012. http://dx.doi.org/10.1016/j.molcel.2011.11.029
- S.L. Bumgarner, R.D. Dowell, P. Grisafi, D.K. Gifford, and G.R. Fink, "Toggle involving cis -interfering noncoding RNAs controls variegated gene expression in yeast", Proceedings of the National Academy of Sciences, vol. 106, pp. 18321-18326, 2009. http://dx.doi.org/10.1073/pnas.0909641106
- D. Holmes, A. Lancaster, S. Lindquist, and R. Halfmann, "Heritable Remodeling of Yeast Multicellularity by an Environmentally Responsive Prion", Cell, vol. 153, pp. 153-165, 2013. http://dx.doi.org/10.1016/j.cell.2013.02.026
- A. Lardenois, I. Stuparevic, Y. Liu, M.J. Law, E. Becker, F. Smagulova, K. Waern, M. Guilleux, J. Horecka, A. Chu, C. Kervarrec, R. Strich, M. Snyder, R.W. Davis, L.M. Steinmetz, and M. Primig, "The conserved histone deacetylase Rpd3 and its DNA binding subunit Ume6 control dynamic transcript architecture during mitotic growth and meiotic development", Nucleic Acids Research, vol. 43, pp. 115-128, 2014. http://dx.doi.org/10.1093/nar/gku1185
- T. Krishnamoorthy, X. Chen, J. Govin, W.L. Cheung, J. Dorsey, K. Schindler, E. Winter, C.D. Allis, V. Guacci, S. Khochbin, M.T. Fuller, and S.L. Berger, "Phosphorylation of histone H4 Ser1 regulates sporulation in yeast and is conserved in fly and mouse spermatogenesis", Genes & Development, vol. 20, pp. 2580-2592, 2006. http://dx.doi.org/10.1101/gad.1457006
- M. Xu, M. Soloveychik, M. Ranger, M. Schertzberg, Z. Shah, R. Raisner, S. Venkatasubrahmanyan, K. Tsui, M. Gebbia, T. Hughes, H. van Bakel, C. Nislow, H. Madhani, and M. Meneghini, "Timing of Transcriptional Quiescence during Gametogenesis Is Controlled by Global Histone H3K4 Demethylation", Developmental Cell, vol. 23, pp. 1059-1071, 2012. http://dx.doi.org/10.1016/j.devcel.2012.10.005
- J.N. McKnight, and T. Tsukiyama, "The conserved HDAC Rpd3 drives transcriptional quiescence in S. cerevisiae", Genomics Data, vol. 6, pp. 245-248, 2015. http://dx.doi.org/10.1016/j.gdata.2015.10.008
- M. Radonjic, J. Andrau, P. Lijnzaad, P. Kemmeren, T.T. Kockelkorn, D. van Leenen, N.L. van Berkum, and F.C. Holstege, "Genome-Wide Analyses Reveal RNA Polymerase II Located Upstream of Genes Poised for Rapid Response upon S. cerevisiae Stationary Phase Exit", Molecular Cell, vol. 18, pp. 171-183, 2005. http://dx.doi.org/10.1016/j.molcel.2005.03.010
- A.T. GANESAN, H. HOLTER, and C. ROBERTS, "Some observations on sporulation in Saccharomyces.", Comptes rendus des travaux du Laboratoire Carlsberg. Serie chimique, 1958. http://www.ncbi.nlm.nih.gov/pubmed/13639529
- B.H. Kirsop, "STUDIES IN YEAST SPORULATION: I. SOME FACTORS INFLUENCING SPORULATION", Journal of the Institute of Brewing, vol. 60, pp. 393-399, 1954. http://dx.doi.org/10.1002/j.2050-0416.1954.tb02767.x
- D. Tsuchiya, Y. Yang, and S. Lacefield, "Positive Feedback of NDT80 Expression Ensures Irreversible Meiotic Commitment in Budding Yeast", PLoS Genetics, vol. 10, pp. e1004398, 2014. http://dx.doi.org/10.1371/journal.pgen.1004398
- E. Winter, "The Sum1/Ndt80 Transcriptional Switch and Commitment to Meiosis in Saccharomyces cerevisiae", Microbiology and Molecular Biology Reviews, vol. 76, pp. 1-15, 2012. http://dx.doi.org/10.1128/MMBR.05010-11
- D. Corbi, S. Sunder, M. Weinreich, A. Skokotas, E.S. Johnson, and E. Winter, "Multisite Phosphorylation of the Sum1 Transcriptional Repressor by S-Phase Kinases Controls Exit from Meiotic Prophase in Yeast", Molecular and Cellular Biology, vol. 34, pp. 2249-2263, 2014. http://dx.doi.org/10.1128/MCB.01413-13
- L. Berchowitz, G. Kabachinski, M. Walker, T. Carlile, W. Gilbert, T. Schwartz, and A. Amon, "Regulated Formation of an Amyloid-like Translational Repressor Governs Gametogenesis", Cell, vol. 163, pp. 406-418, 2015. http://dx.doi.org/10.1016/j.cell.2015.08.060
- L. Jin, K. Zhang, Y. Xu, R. Sternglanz, and A.M. Neiman, "Sequestration of mRNAs Modulates the Timing of Translation during Meiosis in Budding Yeast", Molecular and Cellular Biology, vol. 35, pp. 3448-3458, 2015. http://dx.doi.org/10.1128/MCB.00189-15
- S.M. Honigberg, C. Conicella, and R.E. Espositio, "Commitment to meiosis in Saccharomyces cerevisiae: involvement of the SPO14 gene.", Genetics, 1992. http://www.ncbi.nlm.nih.gov/pubmed/1582554
- S.M. Honigberg, and R.E. Esposito, "Reversal of cell determination in yeast meiosis: postcommitment arrest allows return to mitotic growth.", Proceedings of the National Academy of Sciences, vol. 91, pp. 6559-6563, 1994. http://dx.doi.org/10.1073/pnas.91.14.6559
- H. Yang, Q. Ren, and Z. Zhang, "Chromosome or chromatin condensation leads to meiosis or apoptosis in stationary yeast (Saccharomyces cerevisiae) cells", FEMS Yeast Research, vol. 6, pp. 1254-1263, 2006. http://dx.doi.org/10.1111/j.1567-1364.2006.00123.x
- G.S. Davidson, R.M. Joe, S. Roy, O. Meirelles, C.P. Allen, M.R. Wilson, P.H. Tapia, E.E. Manzanilla, A.E. Dodson, S. Chakraborty, M. Carter, S. Young, B. Edwards, L. Sklar, and M. Werner-Washburne, "The proteomics of quiescent and nonquiescent cell differentiation in yeast stationary-phase cultures", Molecular Biology of the Cell, vol. 22, pp. 988-998, 2011. http://dx.doi.org/10.1091/mbc.E10-06-0499
- C. Allen, S. Büttner, A.D. Aragon, J.A. Thomas, O. Meirelles, J.E. Jaetao, D. Benn, S.W. Ruby, M. Veenhuis, F. Madeo, and M. Werner-Washburne, "Isolation of quiescent and nonquiescent cells from yeast stationary-phase cultures", The Journal of Cell Biology, vol. 174, pp. 89-100, 2006. http://dx.doi.org/10.1083/jcb.200604072
- Z. Palková, and L. Váchová, "Yeast cell differentiation: Lessons from pathogenic and non-pathogenic yeasts", Seminars in Cell & Developmental Biology, vol. 57, pp. 110-119, 2016. http://dx.doi.org/10.1016/j.semcdb.2016.04.006
- S.M. Honigberg, "Cell Signals, Cell Contacts, and the Organization of Yeast Communities", Eukaryotic Cell, vol. 10, pp. 466-473, 2011. http://dx.doi.org/10.1128/EC.00313-10
- S. Piccirillo, and S.M. Honigberg, "Yeast Colony Embedding Method", Journal of Visualized Experiments, 2011. http://dx.doi.org/10.3791/2510
- L. Váchová, L. Hatáková, M. Čáp, M. Pokorná, and Z. Palková, "Rapidly Developing Yeast Microcolonies Differentiate in a Similar Way to Aging Giant Colonies", Oxidative Medicine and Cellular Longevity, vol. 2013, pp. 1-9, 2013. http://dx.doi.org/10.1155/2013/102485
- M. Čáp, L. Štěpánek, K. Harant, L. Váchová, and Z. Palková, "Cell Differentiation within a Yeast Colony: Metabolic and Regulatory Parallels with a Tumor-Affected Organism", Molecular Cell, vol. 46, pp. 436-448, 2012. http://dx.doi.org/10.1016/j.molcel.2012.04.001
- G.S. BAILLIE, and L.J. DOUGLAS, "Role of dimorphism in the development of Candida albicans biofilms", Journal of Medical Microbiology, vol. 48, pp. 671-679, 1999. http://dx.doi.org/10.1099/00222615-48-7-671
- J. Chandra, D.M. Kuhn, P.K. Mukherjee, L.L. Hoyer, T. McCormick, and M.A. Ghannoum, "Biofilm Formation by the Fungal Pathogen Candida albicans : Development, Architecture, and Drug Resistance", Journal of Bacteriology, vol. 183, pp. 5385-5394, 2001. http://dx.doi.org/10.1128/jb.183.18.5385-5394.2001
- L. Váchová, and Z. Palková, "Physiological regulation of yeast cell death in multicellular colonies is triggered by ammonia", The Journal of Cell Biology, vol. 169, pp. 711-717, 2005. http://dx.doi.org/10.1083/jcb.200410064
- L. Váchová, and Z. Palková, "Aging and longevity of yeast colony populations: metabolic adaptation and differentiation", Biochemical Society Transactions, vol. 39, pp. 1471-1475, 2011. http://dx.doi.org/10.1042/BST0391471
- H. Chen, and G.R. Fink, "Feedback control of morphogenesis in fungi by aromatic alcohols", Genes & Development, vol. 20, pp. 1150-1161, 2006. http://dx.doi.org/10.1101/gad.1411806
- L. Váchová, H. Kučerová, F. Devaux, M. Úlehlová, and Z. Palková, "Metabolic diversification of cells during the development of yeast colonies", Environmental Microbiology, vol. 11, pp. 494-504, 2009. http://dx.doi.org/10.1111/j.1462-2920.2008.01789.x
- L. Váchová, O. Chernyavskiy, D. Strachotová, P. Bianchini, Z. Burdíková, I. Ferčíková, L. Kubínová, and Z. Palková, "Architecture of developing multicellular yeast colony: spatio‐temporal expression of Ato1p ammonium exporter", Environmental Microbiology, vol. 11, pp. 1866-1877, 2009. http://dx.doi.org/10.1111/j.1462-2920.2009.01911.x
- Z. Palková, F. Devaux, M. R̆ic̆icová, L. Mináriková, S. Le Crom, and C. Jacq, "Ammonia Pulses and Metabolic Oscillations Guide Yeast Colony Development", Molecular Biology of the Cell, vol. 13, pp. 3901-3914, 2002. http://dx.doi.org/10.1091/mbc.e01-12-0149
- Z. Palková, and J. Forstová, "Yeast colonies synchronise their growth and development.", Journal of cell science, 2000. http://www.ncbi.nlm.nih.gov/pubmed/10806103
- D.M. Gordon, "The rewards of restraint in the collective regulation of foraging by harvester ant colonies", Nature, vol. 498, pp. 91-93, 2013. http://dx.doi.org/10.1038/nature12137
- E.O. Wilson, and B. Hölldobler, "Dense heterarchies and mass communication as the basis of organization in ant colonies", Trends in Ecology & Evolution, vol. 3, pp. 65-68, 1988. http://dx.doi.org/10.1016/0169-5347(88)90018-3
- M. Čáp, L. Váchová, and Z. Palková, "Reactive Oxygen Species in the Signaling and Adaptation of Multicellular Microbial Communities", Oxidative Medicine and Cellular Longevity, vol. 2012, pp. 1-13, 2012. http://dx.doi.org/10.1155/2012/976753
- M. Čáp, L. Váchová, and Z. Palková, "Yeast Colony Survival Depends on Metabolic Adaptation and Cell Differentiation Rather Than on Stress Defense", Journal of Biological Chemistry, vol. 284, pp. 32572-32581, 2009. http://dx.doi.org/10.1074/jbc.M109.022871
- D. St Johnston, and B. Sanson, "Epithelial polarity and morphogenesis", Current Opinion in Cell Biology, vol. 23, pp. 540-546, 2011. http://dx.doi.org/10.1016/j.ceb.2011.07.005
- K.D. Irvine, and C. Rauskolb, "Boundaries in Development: Formation and Function", Annual Review of Cell and Developmental Biology, vol. 17, pp. 189-214, 2001. http://dx.doi.org/10.1146/annurev.cellbio.17.1.189
- A. Rokas, "The Origins of Multicellularity and the Early History of the Genetic Toolkit For Animal Development", Annual Review of Genetics, vol. 42, pp. 235-251, 2008. http://dx.doi.org/10.1146/annurev.genet.42.110807.091513
- L. Hall-Stoodley, J.W. Costerton, and P. Stoodley, "Bacterial biofilms: from the Natural environment to infectious diseases", Nature Reviews Microbiology, vol. 2, pp. 95-108, 2004. http://dx.doi.org/10.1038/nrmicro821
- K.J. Niklas, and S.A. Newman, "The origins of multicellular organisms", Evolution & Development, vol. 15, pp. 41-52, 2013. http://dx.doi.org/10.1111/ede.12013
- A.H. Knoll, "The Multiple Origins of Complex Multicellularity", Annual Review of Earth and Planetary Sciences, vol. 39, pp. 217-239, 2011. http://dx.doi.org/10.1146/annurev.earth.031208.100209
ACKNOWLEDGMENTS
Preparation of this review was supported by funding from the National Institutes of General Medical Sciences of the NIH under award number R15GM094770. I thank Dr. David Spade (Univ. Missouri-Kansas City, UMKC) and Dr. Deendayal Dinakarpandian (UMKC) for helpful discussions, and I am grateful to Dr. Dinakarpandian for preparing Fig. 2C.
COPYRIGHT
© 2016

Similar environments but diverse fates: Responses of budding yeast to nutrient deprivation. by Saul M. Honigberg is licensed under a Creative Commons Attribution 4.0 International License.