Research Articles:
Microbial Cell, Vol. 8, No. 1, pp. 17 - 27; doi: 10.15698/mic2021.01.740
Nutrient sensing and cAMP signaling in yeast: G-protein coupled receptor versus transceptor activation of PKA
1 Laboratory of Molecular Cell Biology, Institute of Botany and Microbiology, KU Leuven, B-3001 Leuven-Heverlee, Flanders, Belgium.
2 Center for Microbiology, VIB, Kasteelpark Arenberg 31, B-3001 Leuven-Heverlee, Flanders, Belgium.
† These authors made an equal contribution to this work.
Keywords: FRET biosensor, cAMP, PKA, nutrient sensing, GPCR, transceptor, yeast.
Abbreviations:
cAMP – cyclic adenosine monophosphate,
EPAC – exchange protein activated by cAMP,
FRET – fluorescence or förster resonance energy transfer,
GPCR – G-protein coupled receptor,
GST – Glutathion-S-Transferase,
PKA – protein kinase A.
Received originally: 17/12/2019 Received in revised form: 12/09/2020
Accepted: 17/09/2020
Published: 12/10/2020
Correspondence:
Johan M. Thevelein, Tel: +32 16 32 15 07; johan.thevelein@kuleuven.vib.be; johan.thevelein@novelyeast.com
Conflict of interest statement: The authors report no conflicts of interest.
Please cite this article as: Griet Van Zeebroeck, Liesbeth Demuyser, Zhiqiang Zhang, Ines Cottignie, Johan M. Thevelein (2020). Nutrient sensing and cAMP signaling in yeast: G-protein coupled receptor versus transceptor activation of PKA. Microbial Cell 8(1): 17-27. doi: 10.15698/mic2021.01.740
Abstract
A major signal transduction pathway regulating cell growth and many associated physiological properties as a function of nutrient availability in the yeast Saccharomyces cerevisiae is the protein kinase A (PKA) pathway. Glucose activation of PKA is mediated by G-protein coupled receptor (GPCR) Gpr1, and secondary messenger cAMP. Other nutrients, including nitrogen, phosphate and sulfate, activate PKA in accordingly-starved cells through nutrient transceptors, but apparently without cAMP signaling. We have now used an optimized EPAC-based fluorescence resonance energy transfer (FRET) sensor to precisely monitor in vivo cAMP levels after nutrient addition. We show that GPCR-mediated glucose activation of PKA is correlated with a rapid transient increase in the cAMP level in vivo, whereas nutrient transceptor-mediated activation by nitrogen, phosphate or sulfate, is not associated with any significant increase in cAMP in vivo. We also demonstrate direct physical interaction between the Gap1 amino acid transceptor and the catalytic subunits of PKA, Tpk1, 2 and 3. In addition, we reveal a conserved consensus motif in the nutrient transceptors that is also present in Bcy1, the regulatory subunit of PKA. This suggests that nutrient transceptor activation of PKA may be mediated by direct release of bound PKA catalytic subunits, triggered by the conformational changes occurring during transport of the substrate by the transceptor. Our results support a model in which nutrient transceptors are evolutionary ancestors of GPCRs, employing a more primitive direct signaling mechanism compared to the indirect cAMP second-messenger signaling mechanism used by GPCRs for activation of PKA.
INTRODUCTION
For all living organisms, but especially for microorganisms, nutrients are a major determinant for cell growth and survival. Any change in nutrient availability and composition in the extracellular environment triggers rapid adaptation mechanisms within the cells to properly adjust cellular properties. With respect to the yeast Saccharomyces cerevisiae, the presence of fermentable sugar in the extracellular medium has major effects on cell growth and physiology. The cells suppress respiration in favor of ethanol fermentation and show rapid growth, characterized by low levels of reserve carbohydrates, low stress resistance, low expression of stress response element (STRE)-controlled genes, weak cell walls and high expression of ribosomal protein genes. On the other hand, when yeast grows on non-fermentable carbon sources, they proliferate more slowly and show the opposite phenotypes of cells growing on fermentable sugar [1]. Addition of glucose or sucrose to these cells activates Protein Kinase A (PKA) through stimulation of cAMP (cyclic adenosine monophosphate) synthesis, mediated by the G-protein coupled receptor (GPCR), Gpr1, and fructose-1,6-bisphosphate activation of Cdc25/Ras, triggering activation of adenylate cyclase [2][3]. However, to maintain the high PKA phenotype typical for fermenting cells, a complete fermentable growth medium is required. When fermenting cells are starved for any other essential nutrient, they will arrest in the G1 phase of the cell cycle, enter the stationary phase G0 and acquire a low PKA phenotype, similar to that of cells proliferating slowly on non-fermentable carbon sources [1].
–
Re-addition of the missing essential nutrient to fermenting stationary-phase cells starved for this nutrient will trigger a similar rapid activation of PKA as addition of a fermentable sugar to respiring cells, and support the re-initiation of rapid fermentable growth. When this upshift is triggered by amino acids, ammonium, phosphate, sulfate, iron or zinc, resupplemented to accordingly-starved fermenting cells, it is not associated with an increase in the cAMP level in spite of the apparent rapid activation of PKA, as inferred from the rapid phosphorylation and adaptation of PKA target systems [4][5][6]. In this case, no evidence has been found for involvement of nutrient-sensing GPCRs, but rather for high-affinity starvation-induced transporters acting as receptors, or transceptors [7][8]. This may be consistent with a more primitive activation mechanism of PKA by nutrient transceptors, preceding in evolution the more elaborate mechanisms employed by dedicated receptors and making use of second-messenger signaling [8][9]. A convenient read-out for rapid activation of PKA is the NTH1-encoded neutral trehalase, which is about tenfold activated by direct PKA phosphorylation, and responsible for the rapid mobilization of trehalose during the initiation or stimulation of fermentation [10].
–
Up to now, all cAMP measurements in the previous studies on nutrient signaling have been performed in vitro using cell extracts obtained after rapid filtering, freezing and subzero destruction of enzyme activity with perchloric acid, and using a commercial kit for cAMP determination in the neutralized extracts [11]. Because of the short time courses, the necessity of subzero elimination of cAMP phosphodiesterase activity and the presence of background signals with some commercial cAMP determination kits, these in vitro measurements have always remained challenging. More recently, Fluorescence or Förster resonance energy transfer (FRET) based methodologies for real-time in vivo monitoring of metabolite levels with excellent temporal resolution have been developed. In 2004, a biosensor was created to monitor levels of cAMP in mammalian cells by tagging the N- and C-terminal ends of the cAMP-binding EPAC (exchange protein activated by cAMP) protein with a FRET donor and acceptor [12][13]. This system has subsequently been optimized further [14][15][16]. In 2017, the first functional S. cerevisiae variant of the EPAC sensor was developed, based on the original EYFP-EPAC2-ECFP construct of Nikolaev et al.[17]. Lately, the research group of Bas Teusink has developed an alternative cAMP sensor for S. cerevisiae based on the mammalian sensor of Klarenbeek et al.[16], using mTurquoise2del as FRET donor and tdTomato as FRET acceptor [18]. The major advantage of this latest sensor is the fact that the fluorescent proteins used are less pH sensitive, within the physiological intracellular pH-range of the yeast cells, enabling these sensors to report the cAMP concentration more reliably and independent of changes in intracellular pH. Since we are studying nutrient-induced effects known to affect intracellular pH, such as the transient glucose-induced drop in intracellular pH [19][20][21], this is an important advantage in experiments aimed at measuring in vivo changes in the cAMP level.
–
We have now employed this optimized cAMP sensor for determination of cAMP levels in vivo during activation of PKA with different nutrients in yeast. We show that glucose triggers rapid cAMP signaling dependent on the GPCR, Gpr1. On the other hand, nutrient transceptor-dependent activation of PKA with nitrogen (amino acids or ammonium), phosphate or sulfate, in accordingly-starved cells is not associated with any significant increase in cAMP. This is consistent with two different signaling mechanisms for nutrient activation of PKA, either GPCR-mediated cAMP signaling or transceptor-mediated cAMP-independent signaling. Moreover, we show that the Gap1 amino acid transceptor interacts directly with the catalytic subunits (Tpk1-3) of PKA, using an in vitro pull-down assay, and that nutrient transceptors contain a conserved consensus sequence, which is also present in the regulatory subunit (Bcy1). These results suggest that transceptors may use a more primitive, direct mechanism for activation of PKA.
RESULTS AND DISCUSSION
Glucose activation of PKA is correlated with cAMP signaling
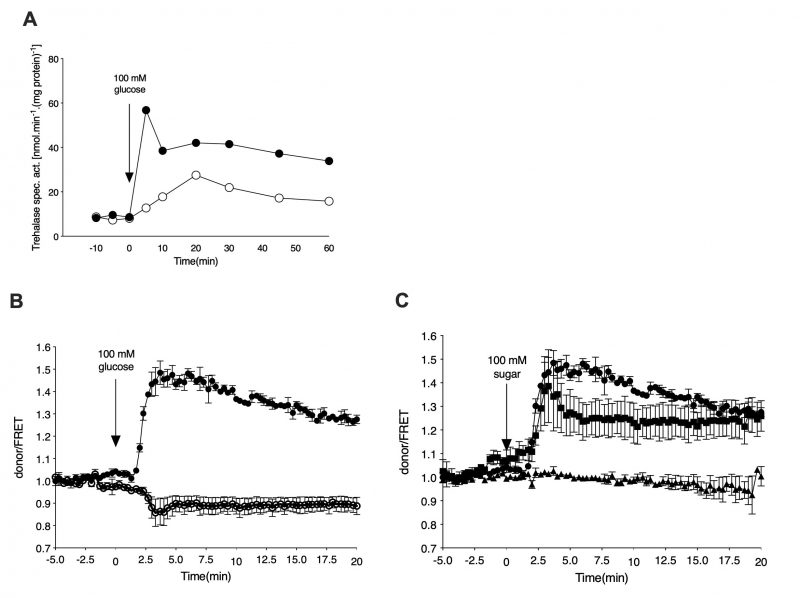
In order to monitor cAMP levels upon nutrient addition in a temporally-resolved manner, we have used the EPAC cAMP sensor as optimized by the lab of Bas Teusink [18]. Yeast cells expressing the cAMP sensor were grown in the absence of glucose, using glycerol as the sole carbon source. After 16 to 24 h of growth, the cells were resuspended in fresh medium and either loaded in the microfluidic device or processed further for trehalase activity determination. Addition of 100 mM glucose to glycerol-grown cells caused a rapid increase in trehalase activity also in the cells expressing the cAMP EPAC sensor (Figure 1A). This increase was dependent on the GPCR, Gpr1, as shown with the gpr1Δ strain (Figure 1A). Using the EPAC sensor, we measured a concomitant increase in the cAMP level upon addition of glucose to glycerol-grown cells (Figure 1B). Deletion of GPR1 fully eliminated this cAMP increase (Figure 1B). These results are consistent with the previous reports of Gpr1-mediated cAMP signaling being responsible for rapid activation of the PKA target trehalase [22][23]. Determination of cAMP levels in vivo with the EPAC sensor after addition of different sugars showed that glucose and sucrose, but not fructose, triggered a rapid increase in cAMP (Figure 1C). This is also consistent with the previously reported sugar specificity of Gpr1 [24].
–
–
Nitrogen, phosphate and sulfate activation of PKA are not correlated with cAMP signaling
In the case of nitrogen starvation, yeast cells expressing the cAMP EPAC sensor were incubated for 24h in the absence of any nitrogen source. The nitrogen-starved cells were resuspended in fresh starvation medium and either loaded in the microfluidic device or processed further for determination of trehalase activation by addition of a nitrogen source. Addition of 5 mM of L-citrulline to nitrogen-starved cells triggers a pronounced activation of trehalase also in the cells expressing the cAMP EPAC sensor, and to a similar extent as seen for glucose addition to respiring cells [25] (Figure 2A). Moreover, the amino acid transceptor Gap1 is essential for this effect as no increase in trehalase activity is observed in the absence of this protein. As opposed to what is observed for glucose, addition of L-citrulline to nitrogen-starved cells does not trigger any increase in the cAMP level in vivo, confirming that the nitrogen source must cause activation of PKA in a cAMP-independent manner (Figure 2B). Similarly, addition of ammonium to nitrogen-starved cells causes a rapid increase in trehalase activity also in the cells expressing the cAMP EPAC sensor (Figure 2C). Absence of trehalase activation in a mep2Δ strain, with Mep2 being the main transceptor for ammonium in S. cerevisiae, confirms nutrient specificity [26] (Figure 2C). As observed with L-citrulline, the increase in trehalase activity upon re-addition of 5 mM ammonium to nitrogen-starved wild type cells, is not correlated with any increase in the cAMP level (Figure 2D). The slight downward trend is possibly due to a technical artefact, such as a focus shift during imaging, rather than reflecting a true change in cAMP levels in vivo.
–
–
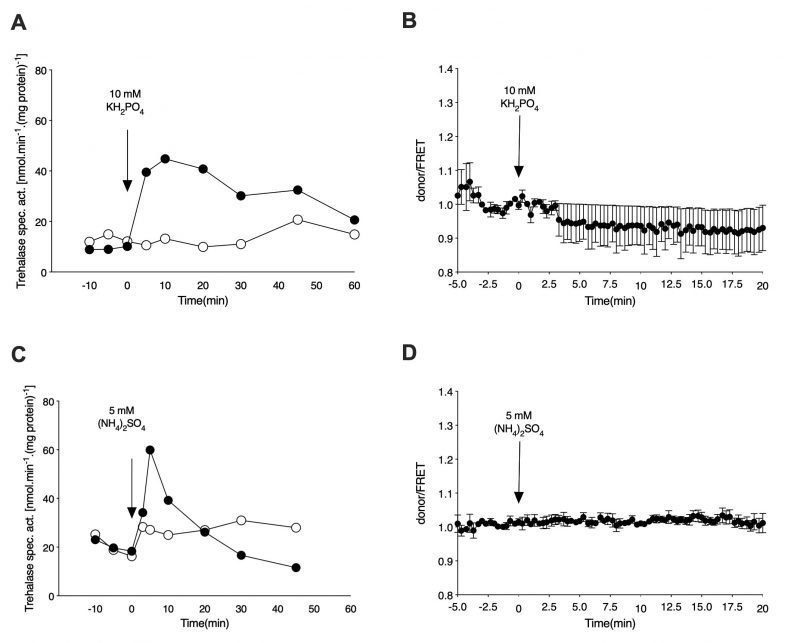
For determination of the cAMP level in vivo upon re-addition of phosphate to phosphate-starved cells, the wild type strain expressing the cAMP EPAC sensor was incubated in phosphate starvation medium for 72 h, with daily refreshment of the medium. As in the case of nitrogen-starved cells, the phosphate-starved cells were washed, resuspended in fresh starvation medium and either loaded in the microfluidic device or used for determination of trehalase activation. Addition of 10 mM KH2PO4 to phosphate-starved cells triggers a rapid increase in the level of trehalase activity also in the cells expressing the cAMP EPAC sensor [27] (Figure 3A). The presence of the phosphate transceptor Pho84 is required for this activation as the pho84Δ strain does not show any increase in activity upon re-addition of phosphate. Despite the pronounced activation of trehalase, which is known to be mediated by PKA [27][28], there is no increase in the level of the secondary messenger cAMP, as inferred from the FRET signal, after addition of phosphate to the phosphate-starved cells (Figure 3B).
–
–
Finally, for determination of the cAMP level in vivo upon re-addition of sulfate to sulfate-starved cells, cells expressing the cAMP EPAC sensor were incubated in sulfate starvation medium for 48 h with daily refreshment of the medium. As observed for nitrogen and phosphate addition, the rapid increase in trehalase activity upon re-addition of sulfate (NH4)2SO4) to sulfur-starved cells was not correlated with an increase in the cAMP level in vivo, as determined by the FRET sensor (Figure 3C and 3D). As shown previously, absence of the increase in trehalase activity in the absence of the sulfate transceptors Sul1 and Sul2 indicates that these proteins mediate activation of PKA upon sulfate addition to sulfur-starved cells [5].
–
Gap1 interacts directly with the catalytic subunits of PKA
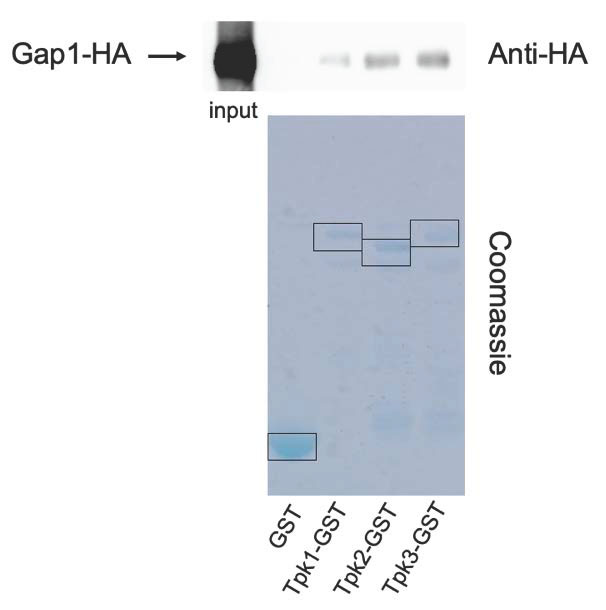
The FRET measurements of in vivo cAMP levels during transceptor activation of PKA, suggest that transceptors activate PKA independent of signaling by the classical second messenger of the GPCR-adenylate cyclase pathway. Hence, an alternative activation mechanism of PKA appears to be involved. We explored the possibility that the transceptors would directly interact with PKA to trigger its activation. For that purpose, we have tested in vitro interaction of the amino acid transceptor Gap1 with the catalytic subunits of PKA, Tpk1, 2 and 3. HA-tagged Gap1 was expressed in nitrogen-starved yeast cells from a plasmid under its own promotor. The Glutathion-S-transferase (GST) tag as such (as a negative control) and GST-tagged Tpk1, 2 or 3 were expressed in Escherichia coli. Gap1-Tpk1, -Tpk2 or -Tpk3 interactions were subsequently tested separately with a GST-pull down assay, in which the GST-tagged catalytic subunits of PKA were precipitated. Co-precipitated HA-tagged Gap1 was detected by Western blot with an anti-HA-HRP antibody. A clear signal of precipitated HA-Gap1 was observed with all three Tpk proteins (Figure 4), indicating that Gap1 binds with Tpk1, 2 and 3 in vitro (Figure 4), while no signal was present for the negative control.
–
–
The physical interaction between Gap1 and the catalytic subunits of PKA suggests a simple mechanism for nutrient transceptor activation of PKA. Sequestration of part of the catalytic subunits by the nutrient transceptors, when they are strongly induced during starvation for their substrate, may help to downregulate PKA activity in the nutrient-starved cells. Stationary phase in yeast is generally correlated with a range of physiological properties indicating low PKA activity [1]. Re-addition of the missing nutrient allows its transport by the transceptor and the associated conformational changes in the transceptor may release the catalytic subunits of PKA, resulting in the typical burst in PKA activity, as indicated by the rapid changes in PKA controlled properties, like the activity of trehalase. This may explain why the activation of trehalase in general is closely correlated with the transport activity of the transceptor [8][25][29].
–
The nutrient transceptors contain a consensus motif also present in Bcy1
We have found a common motif in the previously identified transceptors Gap1, Pho84, Pho87, Sul1, Sul2, Ftr1, Zrt1, Mep2, Mep1: (I/L/φ)XXXXX(I/L/φ)XXTKXXXXXXφXXφ (Figure 5). The most specific element is the TK motif, which is preceded by a hydrophobic amino acid, preferably leucine or isoleucine, with two random amino acids in between, and this residue is in turn preceded by another hydrophobic amino acid, also preferably leucine or isoleucine, with five amino acid residues in between. The TK motif is followed by two hydrophobic amino acids, with six and two random amino acids in between. The consensus motif is found in the C-terminal tail of the transceptors or in an intracellular loop close to the C-terminus. In the Sul1 and Sul2 transceptors, the consensus motif is predicted to be in an extracellular loop, but the structure prediction for Sul1 and Sul2 is uncertain. This consensus motif is also found in the otherwise totally unrelated protein Bcy1, the regulatory subunit of PKA (Figure 5), which is well known to bind the catalytic subunits Tpk1-3. Hence, the presence of this consensus motif may be indicative of physical interaction between a protein and the catalytic subunits of PKA.
–
–
We also found the consensus motif in a number of other transporters, including the active Hxt transporters of which Hxt1 and Hxt6 are shown as example (Figure 5). This may be indicative for possible interaction with PKA and transceptor functionality. The consensus sequence also appears to be present in Cdc33, which is the translation initiation factor eIF4E and mRNA cap binding protein. The conditional cdc33 mutant arrests at the restrictive temperature at the nutrient starvation site in the G1 phase of the cell cycle, just like the conditional mutants cdc25 and cdc35, which are deficient in cAMP-dependent PKA activity at the restrictive temperature [30][31]. The connection between Cdc33 and the cAMP-PKA pathway remained unclear. The presence of the consensus sequence in Cdc33 may indicate that it interacts with the catalytic subunits of PKA and that its inactivation may compromise PKA activity.
–
The consensus motif is not present in the amino acid transporters most closely related to Gap1 and also not in the transporters most closely related to Pho84 (except for Gal2 and Snf3) (Figure 5). This indicates that the consensus motif is not just a common structural or regulatory element in these transporter families. The consensus sequence is also found in the Gal2 galactose/glucose permease and in Snf3, a transporter-like glucose sensor. Connections between the Snf3/Rgt2 glucose sensing pathway and the PKA pathway have been described [32][33].
–
The current data seem to support the concept that the consensus motif identified in this work is not just a structural or regulatory element conserved in a family of transport proteins but that it points to a specific regulatory function that may be related to the specific transceptor functionality in sensing and/or signaling the transport of the nutrient by the transceptor. The presence of the consensus motif in the Bcy1 regulatory subunit of PKA (Figure 5) and the physical binding of the catalytic subunits to the Gap1 transceptor (Figure 4) are consistent with a function of the consensus motif in the interaction with the catalytic subunits Tpk1-3 of PKA. Further research will have to reveal the precise function of this consensus motif.
–
Conclusions
We have shown that the optimized EPAC sensor is a convenient tool to measure rapid changes in the cAMP level in vivo as a function of time in response to the addition of different nutrients. Using this sensor, we confirm that GPCR-mediated glucose activation of PKA in yeast is correlated with a sharp increase in the level of cAMP as second messenger while transceptor-mediated activation of PKA by other nutrients, including nitrogen, phosphate and sulfate, is not correlated with any increase in the cAMP level in vivo. The identification of a consensus motif in the nutrient transceptors that is also present in the Bcy1 regulatory subunit of PKA, and the physical interaction in vitro between the Gap1 transceptor and the Tpk1-3 catalytic subunits of PKA, suggest that nutrient transceptor activation of PKA may be mediated by direct release of bound PKA catalytic subunits, triggered by the conformational changes occurring during transport of the substrate by the transceptor. Our results support a model in which nutrient transceptors are evolutionary ancestors of GPCRs, employing a more primitive direct signaling mechanism compared to the indirect cAMP second-messenger signaling mechanism used by GPCRs for activation of PKA.
MATERIALS AND METHODS
Strains, plasmids and growth conditions
–
All strains and plasmids used in this work are summarized in Table 1 and 2, respectively. For FRET measurements, the wild type strain was transformed with the mTurq2del-EPACdDEPCD-tdTomato plasmid (URA3) kindly provided by Bas Teusink (VU Amsterdam) [18]. For the GST-pull down experiments, the gap1Δ strain was transformed with a plasmid expressing HA-tagged Gap1. For all experiments, cells were cultured under shaking at 200 rpm and at 30°C till exponential phase (OD600 1.5-2). Glucose-derepressed cells were grown in YPGly medium containing 2% bacteriological peptone, 1% yeast extract and 2% glycerol. Nutrient-starved fermenting cells were pregrown in minimal medium, containing 0.17% (w/v) Difco yeast nitrogen base without amino acids and without ammonium sulfate, 0.5% (w/v) ammonium sulfate and 2% (w/v) glucose, containing a complete supplement mixture without uracil (SD-Ura) until exponential phase (OD600 1.5-2). They were harvested and resuspended in the appropriate starvation medium containing 4% glucose, and further incubated with shaking at 30°C. Nitrogen-starved cells were obtained by incubation for 24 h in nitrogen starvation medium containing 0.17% (w/v) Difco yeast nitrogen base without amino acids and without ammonium sulfate. Phosphate-starved cells were obtained by incubation for 72 h (with medium refreshment every 24 h) in phosphate starvation medium (0.57% yeast nitrogen base without phosphate). Sulfur-starved cells were obtained by incubation for 48 h (with medium refreshment every 24 h) in sulfur starvation medium as previously described [5].
–
TABLE 1. S. cerevisiae strains used in this work. |
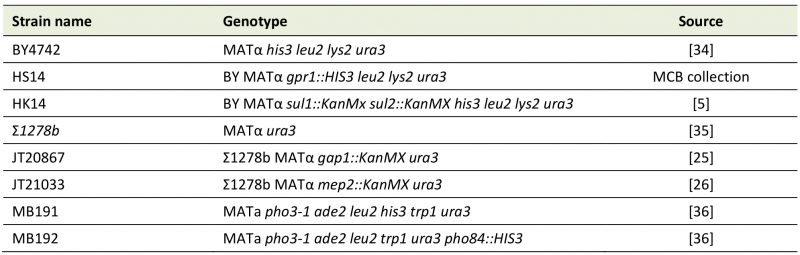 |
| [5][25][26][34][35][36] |
–
Trehalase activity
Trehalase activity as a function of time in response to nutrient addition was determined in cell extracts as previously described [25]. The specific activity of trehalase is expressed as nmol glucose liberated. min-1. (mg protein)-1. Total amount of protein in the samples was determined using the standard Lowry method. Time-course experiments of nutrient-induced trehalase activation were repeated at least three times, representative results are shown for these comparisons between collections of independent data points.
–
TABLE 2. Plasmids used in this work. |
 |
–
Ratiometric FRET measurements
Microfluidics and imaging
–
The CellASIC ONIX2 Microfluidic Platform from Millipore for automated addition of compounds to cells during continuous imaging by microscopy was used, as previously described [37]. The cells were loaded into a chamber of a microwell plate (Y04C, Merck), and liquid was continuously fluxed through the cell chamber. The first well was filled with the appropriate nutrient starvation medium, the second well with starvation medium supplemented with the missing nutrient. The first liquid was fluxed for 10 min, after which we switched to the second liquid for another 20 min. During fluxing, images of the cells in the donor and FRET channels were taken every 20 s. The donor fluorophore was visualized using the 458 nm laser line, excitation DM458/515, SDM560 and emission BA480-495 filter. FRET was monitored using the 458 nm laser line and emission BA575-675 filter. Images were taken by scanning at 8.0 µs/pixel and the 60x UPlanSApo (NA 1.35) objective lens was used in combination with a digital zoom of 1.5x.
–
Data analysis
–
The FRETR ratios were calculated for an average of 20 cells by dividing the corrected acceptor value by the corrected donor value, as previously described [37]. Correction was achieved by subtracting the background and the autofluorescence signal intensity from the sample signal intensity of every cell. In order to visually compare the response of all cells, the values were divided by the average of the corrected values taken a short time period before addition of the compound. For the EPAC sensor, the reverse ratios are shown, since an increase in cAMP results in a decrease in FRET. Mean values of at least three biological replicates, normalised to the value before sugar addition (0 min), are shown with the standard error of the mean.
–
GST-pull down
Expression and purification of GST-tagged proteins from E. coli
–
Proteins were expressed in the E. coli strain BL21 and expression induced by addition of 0.3 mM IPTG (final concentration) and cells were harvested and washed once with ice-cold PBS buffer. Cells were then resuspended in 5 ml of lysis buffer (PBS 1x, 0.4% Triton X-100, 2 mM MgCl2, 1 mM EDTA pH 8.0, 2 mM DTT, 0.2 mg/ml lysozyme and protease inhibitor mix, complete EDTA free, Roche) and incubated on ice for 15 min. Lysis was completed by 3×15 s pulses of sonication. Lysates were clarified by centrifugation for 10 min at 12,000 g at 4°C. The resulting supernatant fraction was incubated with 400 µl of a 50:50 slurry of glutathione sepharose beads (GEHealthcare) (pre-equilibrated in wash buffer (PBS 1x, 0.1% Triton X-100, 2 mM MgCl2, 1 mM EDTA, 1 mM DTT)) in a rollerdrum for 1 h at 4°C. Beads were collected by centrifugation at 500 g for 2 min at 4°C and washed 5 times with wash buffer.
–
Expression and purification of HA-tagged proteins from yeast
–
Cells were harvested and washed once with ice-cold PBS buffer. Cells were resuspended in 500 µl ice-cold lysis buffer (PBS 1x, 0.1% Triton X-100, 10% glycerol, 2.5 mM MgCl2, 1 mM EDTA, 1 mM DTT, 10 mM NaF, 0.4 mM Na3VO4, 0.1 mM b-glycerophosphate, containing protease inhibitor mix, complete EDTA free, Roche). Glass beads were added and cells were lysed by vigorous vortexing (4x1min, with cooling on ice in between). Lysates were clarified by centrifugation at 12,000 g for 10 min at 4°C. Supernatant was transferred to a new microcentrifuge tube and centrifuged for a second time at 12,000 g. Clarified extracts were kept on ice for further use in pull down assays.
–
GST-pull down assay
–
GST fusion proteins were extracted from BL21 E. coli cells as described. Beads were resuspended in 500 µl binding buffer (PBS 1x, 0.05% Triton X-100, 0.1 mM DTT). Yeast cell extracts were prepared as described and clarified extracts were incubated for 60 min at 4°C with 50 µl glutathione sepharose beads (GE Healthcare) to reduce aspecific binding. Beads were collected with a brief spin at 500 g and the resulting supernatant was incubated with equal amounts of beadbound purified GST fusion proteins, prepared as described. After a 2 h incubation at 4°C, samples were allowed to stand for 5 min on ice. The sedimented beads were washed three times with PBS-T (PBS 1x, 0.1% Triton X-100). Finally, proteins were solubilized by adding SDS sample buffer, separated by SDS-PAGE, and visualized by Coomassie staining or immunoblotting with HRP-coupled anti-HA antibody.
REFERENCES
- Thevelein JM, de Winde JH (1999). Novel sensing mechanisms and targets for the cAMP-protein kinase A pathway in the yeast Saccharomyces cerevisiae. Mol Microbiol 33(5): 904-918. org/10.1046/j.1365-2958.1999.01538.x
- Rolland F, De Winde JH, Lemaire K, Boles E, Thevelein JM, Winderickx J (2000). Glucose-induced cAMP signalling in yeast requires both a G-protein coupled receptor system for extracellular glucose detection and a separable hexose kinase-dependent sensing process. Mol Microbiol 38(2): 348-358. org/10.1046/j.1365-2958.2000.02125.x
- Peeters K, Van Leemputte F, Fischer B, Bonini BM, Quezada H, Tsytlonok M, Haesen D, Vanthienen W, Bernardes N, Gonzalez-Blas CB, Janssens V, Tompa P, Versees W, Thevelein JM (2017). Fructose-1,6-bisphosphate couples glycolytic flux to activation of Ras. Nat Commun 8(1): 922. 10.1038/s41467-017-01019-z
- Hirimburegama K, Durnez P, Keleman J, Oris E, Vergauwen R, Mergelsberg H, Thevelein JM (1992). Nutrient-induced activation of trehalase in nutrient-starved cells of the yeast Saccharomyces cerevisiae: cAMP is not involved as second messenger. J Gen Microbiol 138(10): 2035-2043. 10.1099/00221287-138-10-2035
- Kankipati HN, Rubio-Texeira M, Castermans D, Diallinas G, Thevelein JM (2015). Sul1 and Sul2 sulfate transceptors signal to protein kinase A upon exit of sulfur starvation. J Biol Chem 290(16): 10430-10446. 10.1074/jbc.M114.629022
- Schothorst J, Zeebroeck GV, Thevelein JM (2017). Identification of Ftr1 and Zrt1 as iron and zinc micronutrient transceptors for activation of the PKA pathway in Saccharomyces cerevisiae. Microbial Cell 4(3): 74-89. 10.15698/mic2017.03.561
- Holsbeeks I, Lagatie O, Van Nuland A, Van de Velde S, Thevelein JM (2004). The eukaryotic plasma membrane as a nutrient-sensing device. Trends Biochem Sci 29(10): 556-564. 10.1016/j.tibs.2004.08.010
- Steyfkens F, Zhang Z, Van Zeebroeck G, Thevelein JM (2018). Multiple Transceptors for Macro- and Micro-Nutrients Control Diverse Cellular Properties Through the PKA Pathway in Yeast: A Paradigm for the Rapidly Expanding World of Eukaryotic Nutrient Transceptors Up to Those in Human Cells. Front Pharmacol 9: 191. 10.3389/fphar.2018.00191
- Thevelein JM, Voordeckers K (2009). Functioning and evolutionary significance of nutrient transceptors. Mol Biol Evol 26(11): 2407-2414. 10.1093/molbev/msp168
- Schepers W, Van Zeebroeck G, Pinkse M, Verhaert P, Thevelein JM (2012). In vivo phosphorylation of Ser21 and Ser83 during nutrient-induced activation of the yeast protein kinase A (PKA) target trehalase. J Biol Chem 287(53): 44130-44142. 10.1074/jbc.M112.421503
- Thevelein JM, Beullens M, Honshoven F, Hoebeeck G, Detremerie K, den Hollander JA, Jans AW (1987). Regulation of the cAMP level in the yeast Saccharomyces cerevisiae: intracellular pH and the effect of membrane depolarizing compounds. J Gen Microbiol 133(8): 2191-2196. 10.1099/00221287-133-8-2191
- Nikolaev VO, Bunemann M, Hein L, Hannawacker A, Lohse MJ (2004). Novel single chain cAMP sensors for receptor-induced signal propagation. J Biol Chem 279(36): 37215-37218. 10.1074/jbc.C400302200
- Ponsioen B, Zhao J, Riedl J, Zwartkruis F, van der Krogt G, Zaccolo M, Moolenaar WH, Bos JL, Jalink K (2004). Detecting cAMP-induced Epac activation by fluorescence resonance energy transfer: Epac as a novel cAMP indicator. EMBO Rep 5(12): 1176-1180. 10.1038/sj.embor.7400290
- van der Krogt GN, Ogink J, Ponsioen B, Jalink K (2008). A comparison of donor-acceptor pairs for genetically encoded FRET sensors: application to the Epac cAMP sensor as an example. PLoS One 3(4): e1916. 10.1371/journal.pone.0001916
- Klarenbeek JB, Goedhart J, Hink MA, Gadella TW, Jalink K (2011). A mTurquoise-based cAMP sensor for both FLIM and ratiometric read-out has improved dynamic range. PLoS One 6(4): e19170. 10.1371/journal.pone.0019170
- Klarenbeek J, Goedhart J, van Batenburg A, Groenewald D, Jalink K (2015). Fourth-generation epac-based FRET sensors for cAMP feature exceptional brightness, photostability and dynamic range: characterization of dedicated sensors for FLIM, for ratiometry and with high affinity. PLoS One 10(4): e0122513. 10.1371/journal.pone.0122513
- Colombo S, Broggi S, Collini M, D'Alfonso L, Chirico G, Martegani E (2017). Detection of cAMP and of PKA activity in Saccharomyces cerevisiae single cells using Fluorescence Resonance Energy Transfer (FRET) probes. Biochem Biophys Res Commun 487(3): 594-599. 10.1016/j.bbrc.2017.04.097
- Botman D, O'Toole TG, Goedhart J, Bruggeman FJ, van Heerden JH, Teusink B (2019). A new FRET biosensor enlightens cAMP signalling in budding yeast. bioRxiv. 10.1101/831354
- Thevelein JM, Beullens M, Honshoven F, Hoebeeck G, Detremerie K, Griewel B, den Hollander JA, Jans AW (1987). Regulation of the cAMP level in the yeast Saccharomyces cerevisiae: the glucose-induced cAMP signal is not mediated by a transient drop in the intracellular pH. J Gen Microbiol 133(8): 2197-2205. 10.1099/00221287-133-8-2197
- van Heerden JH, Wortel MT, Bruggeman FJ, Heijnen JJ, Bollen YJ, Planque R, Hulshof J, O'Toole TG, Wahl SA, Teusink B (2014). Lost in transition: start-up of glycolysis yields subpopulations of nongrowing cells. Science 343(6174): 1245114. 10.1126/science.1245114
- Van Aelst L, Hohmann S, Bulaya B, de Koning W, Sierkstra L, Neves MJ, Luyten K, Alijo R, Ramos J, Coccetti P, Martegani E, de Magalhães-Rocha NM, Brandão RL, Van Dijck P, Vanhalewyn M, Durnez P, Jans AWH, Thevelein JM (1993). Molecular cloning of a gene involved in glucose sensing in the yeast Saccharomyces cerevisiae. Mol Microbiol 8(5): 927-943. org/10.1111/j.1365-2958.1993.tb01638.x
- Colombo S, Ma P, Cauwenberg L, Winderickx J, Crauwels M, Teunissen A, Nauwelaers D, de Winde JH, Gorwa MF, Colavizza D, Thevelein JM (1998). Involvement of distinct G-proteins, Gpa2 and Ras, in glucose- and intracellular acidification-induced cAMP signalling in the yeast Saccharomyces cerevisiae. EMBO J 17(12): 3326-3341. 10.1093/emboj/17.12.3326
- Kraakman L, Lemaire K, Ma P, Teunissen AW, Donaton MC, Van Dijck P, Winderickx J, de Winde JH, Thevelein JM (1999). A Saccharomyces cerevisiae G-protein coupled receptor, Gpr1, is specifically required for glucose activation of the cAMP pathway during the transition to growth on glucose. Mol Microbiol 32(5): 1002-1012. org/10.1046/j.1365-2958.1999.01413.x
- Lemaire K, Van de Velde S, Van Dijck P, Thevelein JM (2004). Glucose and sucrose act as agonist and mannose as antagonist ligands of the G protein-coupled receptor Gpr1 in the yeast Saccharomyces cerevisiae. Mol Cell 16(2): 293-299. 10.1016/j.molcel.2004.10.004
- Donaton MC, Holsbeeks I, Lagatie O, Van Zeebroeck G, Crauwels M, Winderickx J, Thevelein JM (2003). The Gap1 general amino acid permease acts as an amino acid sensor for activation of protein kinase A targets in the yeast Saccharomyces cerevisiae. Mol Microbiol 50(3): 911-929. org/10.1046/j.1365-2958.2003.03732.x
- Van Nuland A, Vandormael P, Donaton M, Alenquer M, Lourenco A, Quintino E, Versele M, Thevelein JM (2006). Ammonium permease-based sensing mechanism for rapid ammonium activation of the protein kinase A pathway in yeast. Mol Microbiol 59(5): 1485-1505. 10.1111/j.1365-2958.2005.05043.x
- Giots F, Donaton MC, Thevelein JM (2003). Inorganic phosphate is sensed by specific phosphate carriers and acts in concert with glucose as a nutrient signal for activation of the protein kinase A pathway in the yeast Saccharomyces cerevisiae. Mol Microbiol 47(4): 1163-1181. org/10.1046/j.1365-2958.2003.03365.x
- Popova Y, Thayumanavan P, Lonati E, Agrochao M, Thevelein JM (2010). Transport and signaling through the phosphate-binding site of the yeast Pho84 phosphate transceptor. Proc Natl Acad Sci U S A 107(7): 2890-2895. 10.1073/pnas.0906546107
- Samyn DR, Ruiz-Pavon L, Andersson MR, Popova Y, Thevelein JM, Persson BL (2012). Mutational analysis of putative phosphate- and proton-binding sites in the Saccharomyces cerevisiae Pho84 phosphate:H(+) transceptor and its effect on signalling to the PKA and PHO pathways. Biochem J 445(3): 413-422. 10.1042/BJ20112086
- Reed SI (1980). The selection of S. cerevisiae mutants defective in the start event of cell division. Genetics 95(561-577. 7002718
- Pringle JR, Hartwell LH (1981). The Saccharomyces cerevisiae cell cycle. In: Strathern JN, Jones EW, Broach JR, editors. The Molecular Biology of the Yeast Saccharomyces Metabolism and Gene Expression Cold Spring Harbor Laboratory, Cold Spring Harbor; pp 97-142.
- Kim JH, Johnston M (2006). Two glucose-sensing pathways converge on Rgt1 to regulate expression of glucose transporter genes in Saccharomyces cerevisiae. J Biol Chem 281(36): 26144-26149. 10.1074/jbc.M603636200
- Jouandot D, 2nd, Roy A, Kim JH (2011). Functional dissection of the glucose signaling pathways that regulate the yeast glucose transporter gene (HXT) repressor Rgt1. J Cell Biochem 112(11): 3268-3275. 10.1002/jcb.23253
- Brachmann CB, Davies A, Cost GJ, Caputo E, Li J, Hieter P, Boeke JD (1998). Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14(2): 115-132. 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2
- Grenson M (1969). The utilization of exogenous pyrimidines and the recycling of uridine-5'-phosphate derivatives in Saccharomyces cerevisiae, as studied by means of mutants affected in pyrimidine uptake and metabolism. Eur J Biochem 11(2): 249-260. 10.1111/j.1432-1033.1969.tb00767.x
- Bun-Ya M, Nishimura M, Harashima S, Oshima Y (1991). The PHO84 gene of Saccharomyces cerevisiae encodes an inorganic phosphate transporter. Mol Cell Biol 11(6): 3229-3238. 10.1128/mcb.11.6.3229
- Demuyser L, Van Genechten W, Mizuno H, Colombo S, Van Dijck P (2018). Introducing fluorescence resonance energy transfer-based biosensors for the analysis of cAMP-PKA signalling in the fungal pathogen Candida glabrata. Cell Microbiol 20(10): e12863. 10.1111/cmi.12863
–
ACKNOWLEDGMENTS
We thank Ilse Palmans for excellent technical assistance with the experiments and Nico Van-goethem for help with preparation of the figures. This work was supported by a Concerted Research Actions grant (GOA/13/006) and Interuniversity Attraction Poles grant (I.A.P. Network P7/40) to JMT.
COPYRIGHT
© 2020

Nutrient sensing and cAMP signaling in yeast: G-protein coupled receptor versus transceptor activation of PKA by Van Zeebroeck et al. is licensed under a Creative Commons Attribution 4.0 International License.