Research Articles:
Microbial Cell, Vol. 4, No. 10, pp. 342 - 361; doi: 10.15698/mic2017.10.594
Ydj1 governs fungal morphogenesis and stress response, and facilitates mitochondrial protein import via Mas1 and Mas2
1 Aberdeen Fungal Group, University of Aberdeen, Institute of Medical Sciences, Foresterhill, Aberdeen, AB25 2ZD, UK.
2 Department of Molecular Genetics, University of Toronto, Toronto, Ontario, M5S 1A8, Canada.
3 Department of Biochemistry, University of Nebraska, Lincoln, NE 68588, USA.
4 Lunenfeld-Tanenbaum Research Institute, Sinai Health System, 600 University Avenue, Toronto, ON, M5G 1X5, Canada.
5 Nebraska Redox Biology Center, University of Nebraska, Lincoln, NE 68588, USA.
6 Fred & Pamela Buffett Cancer Center, Omaha, NE 68198, USA.
#These authors contributed equally to this work
Keywords: Candida albicans, stress, mitochondria, morphogenesis, heat shock, mitochondrial processing peptidases.
Abbreviations:
MIC - minimum inhibitory concentration,
MPP - mitochondrial processing peptidases,
PMF - post-mitochondrial fraction,
WCE - whole-cell extract.
Received originally: 29/08/2017 Accepted: 13/09/2017
Published: 02/10/2017
Correspondence:
Leah E. Cowen, Department of Molecular Genetics, University of Toronto, Toronto, Ontario, M5S 1A8, Canada; leah.cowen@utoronto.ca
Conflict of interest statement: The authors declare no conflict of interest.
Please cite this article as: Jinglin L. Xie, Iryna Bohovych, Erin O.Y. Wong, Jean-Philippe Lam-bert, Anne-Claude Gingras, Oleh Khalimonchuk, Leah E. Cowen and Michelle D. Leach. Ydj1 governs fungal morphogenesis and stress response, and facilitates mitochondrial protein import via Mas1 and Mas2. Microbial Cell: 4(10): 342-361. doi: 10.15698/mic2017.10.594
Abstract
Mitochondria underpin metabolism, bioenergetics, signalling, development and cell death in eukaryotes. Most of the ~1,000 yeast mitochondrial proteins are encoded in the nucleus and synthesised as precursors in the cytosol, with mitochondrial import facilitated by molecular chaperones. Here, we focus on the Hsp40 chaperone Ydj1 in the fungal pathogen Candida albicans, finding that it is localised to both the cytosol and outer mitochondrial membrane, and is required for cellular stress responses and for filamentation, a key virulence trait. Mapping the Ydj1 protein interaction network highlighted connections with co-chaperones and regulators of filamentation. Furthermore, the mitochondrial processing peptidases Mas1 and Mas2 were highly enriched for interaction with Ydj1. Additional analysis demonstrated that loss of MAS1, MAS2 or YDJ1 perturbs mitochondrial morphology and function. Deletion of YDJ1 impairs import of Su9, a protein that is cleaved to a mature form by Mas1 and Mas2. Thus, we highlight a novel role for Ydj1 in cellular morphogenesis, stress responses, and mitochondrial import in the fungal kingdom.
INTRODUCTION
Mitochondria are essential eukaryotic organelles important for metabolism, bioenergetics, signalling, apoptosis and developmental processes [1]. As a consequence, maintenance of mitochondrial activity by preserving protein localization and function is key to survival. Most mitochondrial proteins are synthesised with signal presequences in the cytosol nd translocated in an unfolded state through the mitochondrial membranes with the help of chaperone proteins [2][3]. In the model yeast Saccharomyces cerevisiae, the signal presequences are proteolytically removed upon their arrival in the mitochondria by the matrix-located mitochondrial processing peptidases (MPP), encoded by MAS1 and MAS2, enabling final protein folding [4][5]. MAS1 and MAS2 were originally identified by isolating temperature-sensitive yeast mutants that accumulated uncleaved mitochondrial precursor proteins at the nonpermissive temperature of 37°C [6]. This screen also identified MAS3 and MAS5, which correspond to the heat shock transcription factor HSF1 and the Hsf1 regulated chaperone YDJ1, respectively [7][8].
–
Ydj1 belongs to the Hsp40 class of chaperones, which are homologous to the DnaJ chaperone in E. coli and interact with the molecular chaperone Hsp70, accelerating its ATPase activity [9]. Deletion of YDJ1 in S. cerevisiae results in slow-growing, stress-sensitive cells, with full Ydj1 function being dependent on farnesylation at the C-terminus [10]. The C-terminal domain of Ydj1 binds substrates with a specificity that overlaps with that of Hsp70 [11], and together Hsp70 and Ydj1 are capable of refolding denatured luciferase [12]. Ydj1 localises to the cytoplasm and endoplasmic reticulum in S. cerevisiae, with a small fraction localising to mitochondria [13]. It was later discovered that a ydj1Δ mutant exhibits defects in mitochondrial import of the nuclear-encoded F1β subunit of the mitochondrial ATPase subunit, accumulating greater levels of the precursor form as the temperature is increased from 30°C to 37°C [8][14]. Although it is clear that Ydj1 influences mitochondrial protein import, the mechanisms involved remain largely enigmatic.
–
Much of our knowledge regarding factors governing proteome stability has been derived by studying stress responses in S. cerevisiae due to its facile genetics. Dissecting these cellular stress responses is of central importance not only to understand fundamental mechanisms of protein homeostasis, but also to appreciate mechanisms of virulence in fungal pathogens. This is of particular relevance for the commensal fungus Candida albicans, which has evolved in the mucous membranes and digestive tracts of healthy humans, causing superficial mucosal infections in otherwise healthy individuals upon compromise of host defences [15]. Moreover, immunocompromised patients are at risk of developing life-threatening systemic infections with mortality rates of ~40% [16]. C. albicans has evolved fine-tuned circuitry to sense and respond to diverse stresses relevant to the human host [17]. It senses temperature and other host cues, which induce a morphological transition between yeast and filamentous growth, a key virulence trait for dissemination and epithelial invasion [18]. Temperature sensing is governed in part by Hsf1 [19], which has crucial functions in orchestrating the expression of genes encoding molecular chaperones involved in basal protein homeostasis such as Ydj1, and the heat shock response [20][21]. There is a growing appreciation of the mechanisms by which Hsf1 and the molecular chaperone Hsp90 govern C. albicans biology [22][23][24][25][26], although the functions of other molecular chaperones regulated by Hsf1 remain a largely uncharted frontier.
–
The importance of mitochondria in virulence, morphogenesis and stress responses in C. albicans has been recently highlighted [27]. Mitochondrial proteins have been linked to cell integrity, being required for tolerance to the antifungal drug caspofungin [28][29], and survival during oxidative stress [30]. Furthermore, loss of mitochondrial proteins blocks filamentation [31][32], which likely accounts for the attenuated virulence of mutants with defective mitochondria [30]. In this study, we investigated the uncharacterised C. albicans Hsp40 chaperone Ydj1. We determined that Ydj1 promotes survival in response to oxidative, cell wall and osmotic stress, and is required for growth at high temperature. In addition, we find that Ydj1 is required for filamentation in response to serum and high temperature cues. Utilising a proteomic approach, we identified numerous Ydj1 interactors, discovering a novel role for a pool of mitochondria-associated Ydj1 in facilitating mitochondrial import through the MPP Mas1 and Mas2. We further showed that this unexpected association is important for maintaining mitochondrial morphology and functionality at elevated temperatures.
RESULTS
Ydj1 is required for stress tolerance and morphogenesis in C. albicans
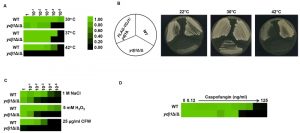
Environmental stress can lead to a breakdown in intracellular protein transport, disrupt the cytoskeleton and trigger global protein unfolding [33], threatening cellular viability. Heat shock proteins are deployed to counteract the detrimental influence of such stressors, preventing protein aggregation and targeting damaged proteins for degradation [34]. To establish if Ydj1 is required for responding to stress in C. albicans, we first determined if Ydj1 influenced thermal adaptation. Wild-type and ydj1Δ/ydj1Δ mutant cells were diluted 10-fold in YPD and placed at 30°C, 37°C or 42°C for 72 hours. The ydj1Δ/ydj1Δ mutant failed to grow at 42°C (Figure 1A), implicating Ydj1 as crucial for growth at elevated temperatures, as is the case in S. cerevisiae [8]. S. cerevisiae Ydj1 is also important for optimal growth at lower temperatures [8], consistent with our observations at 30°C (Figure 1A). To ensure the phenotypes are attributable to deletion of YDJ1, the ydj1Δ/ydj1Δ mutant was complemented with a FLAG-YDJ1 allele at the native locus, and strains were tested for their ability to grow at 22°C, 30°C or 42°C for 48 hours (Figure 1B). The FLAG-YDJ1/ydj1Δ allele partially rescued the slow growth phenotype of the ydj1Δ/ydj1Δ mutant, validating the mutant phenotypes. Next, we examined the effect of osmotic, oxidative and cell wall stress. The ydj1Δ/ydj1Δ mutant was sensitive to osmotic and oxidative stress, and was unable to grow in the presence of the cell wall stress agent calcofluor white (Figure 1C). The striking cell wall phenotype prompted us to test whether a ydj1Δ/ydj1Δ mutant would be hypersensitive to the echinocandin antifungal drug, caspofungin, which exerts a profound cell wall stress in C. albicans [35]. Indeed, ydj1Δ/ydj1Δ mutant cells were hypersensitive to caspofungin compared to the wild-type strain (Figure 1D), suggesting that Ydj1 is required for cell wall integrity.
–
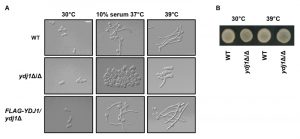
The cell wall of C. albicans is critical for the maintenance of cell polarity and interaction with the surrounding environment. Numerous environmental cues induce a morphogenetic switch from a yeast to filamentous form, during which the expression of cell wall proteins is highly regulated [36]. Given that the ydj1Δ/ydj1Δ mutant is hypersensitive to cell wall stressors, and that other heat shock proteins have been implicated in filamentation [37][38], we tested the ability of a ydj1Δ/ydj1Δ mutant to transition to filamentous growth in response to serum (37°C with 10% serum) or high temperature (39°C) (Figure 2A). The wild type and complemented FLAG-YDJ1/ydj1Δ strain produced elongated hyphae in response to both conditions tested, whereas the ydj1Δ/ydj1Δ mutant was blocked in filamentation. Viability of the ydj1Δ/ydj1Δ mutant was verified after prolonged periods at 39°C by spotting imaged cells onto YPD and incubating at 30°C for 48 hours (Figure 2B). Together, our data demonstrate that Ydj1 is required for optimal growth at 30°C and is essential for high temperature growth. In addition, Ydj1 enables diverse responses to cellular stress and facilitates morphogenesis in response to host-relevant cues.
–
Ydj1 interacts with other co-chaperones and the MPP Mas1 and Mas2
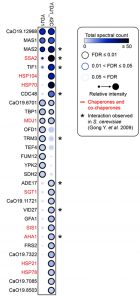
The pleiotropic phenotypes exhibited by the ydj1Δ/ydj1Δ mutant suggest that it interacts with and aids in the folding of numerous proteins. Recent studies in S. cerevisiae identified 64 physical interactions with Ydj1, including 11 chaperone interactors [39][40]. To identify Ydj1 interactors in C. albicans that might influence stress responses, such as heat shock, and morphogenesis, we performed co-immunoprecipitation coupled to mass spectrometry. Wild-type and 2xFLAG-YDJ1/ydj1Δ strains were grown at 30°C or subjected to a 30°C – 42°C heat shock. We identified 30 proteins that interact with Ydj1 in the absence or presence of heat shock, including eight chaperone proteins (Figure 3). The interaction of Ydj1 with additional chaperone proteins was expected based on findings in S. cerevisiae, where Ydj1 interacts with Hsp104 [41], Sis1 and Hsp78 [39], working with Hsp104 and the Hsp70 co-chaperone Ssa1 to support protein refolding [41]. We identified Hsp104, Hsp70, the Hsp70 co-chaperone Ssa2, Sis1, and Hsp78 as interactors of Ydj1 in C. albicans, suggesting that it has a conserved role in protein folding. In addition to these chaperones, we also found Ydj1 to interact with the mitochondrial chaperone Mdj1, the Hsp90 co-chaperone Aha1, and Hsp21 (Figure 3).
–
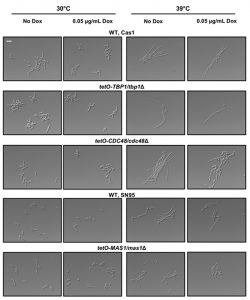
Our observations that the ydj1Δ/ydj1Δ mutant is defective in filamentation suggests that Ydj1 is required for the initiation of filamentous growth. Hsp21 contributes to the formation of filaments [38], but homozygous deletion mutants are not fully blocked in the yeast to filament transition implicating additional factors. Analysis of our Ydj1 interactors highlighted several positive regulators of filamentation in response to serum: Gfa1 [42], Cdc48, Tbp1 and Mas1 [43]. However, these proteins are essential for C. albicans growth, thus it is important to distinguish specific functions in filamentation from confounding effects on viability. We tested the tetO-CDC48/cdc48Δ and tetO-TBP1/tbp1Δ doxycycline-repressible strains from the GRACE (gene replacement and conditional expression) collection [44], along with a tetO-MAS1/mas1Δ strain that we constructed, in conditions that supported viability for morphogenesis in response to elevated temperature of 39°C (Figure 4). In the presence of 0.05 μg/mL doxycycline to repress target gene expression, the tetO-TBP1/tbp1Δ strain filamented robustly, whereas the tetO-CDC48/cdc48Δ and tetO-MAS1/mas1Δ strains displayed a partial defect, with a mixture of yeast and filamentous cells present. The partial defect could be due to stochastic differences in the level of target protein still present in the cell at the point of depletion. Given that both Mas1 and Cdc48 are involved in mitochondrial function in S. cerevisiae [5][45], and that mitochondria play a role in filamentation [32], our data suggests that Yjd1 may promote filamentation in part via proteins important for mitochondrial function.
–
Mas1 and Mas2 are required for mitochondrial function in C. albicans
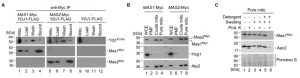
Our most striking interaction detected was with the MPP enzymes Mas1 and Mas2 (Figure 3). They remain uncharacterised in C. albicans, but the orthologues in S. cerevisiae are required for cleaving the N-terminal targeting signal off nuclear encoded mitochondrial proteins upon import [6]. S. cerevisiae cells depleted of one or both MAS subunits continue to import precursor proteins in the mitochondria, but fail to cleave them, leading to cell death [46]. The physical interaction identified between Ydj1 and Mas1/Mas2 by mass spectrometry suggests shared functional relationships. To validate the physical interaction, and ensure that the MPPs did not adventitiously bind to Ydj1 after cell disruption, we performed co-immunoprecipitation from gradient purified mitochondria coupled to Western blot analysis. Immunoprecipitation of Myc-tagged Mas1 or Mas2 with anti-Myc resin co-purified both the Myc-tagged Mas proteins and FLAG-tagged Ydj1 (Figure 5A). For the control strain lacking the tagged MAS alleles, Ydj1 was present in the input (mitochondria) but was not immunoprecipitated (bound fraction) (Figure 5A).
–
To determine if Mas1 and Mas2 are important for mitochondrial function in C. albicans, we first looked at Mas1 and Mas2 localisation. Strains with the C-terminally Myc-tagged Mas1 and Mas2 proteins were grown in YPD for 16 hours, after which whole-cell extracts, post-mitochondrial fractions, crude and gradient purified (pure) mitochondria were extracted and analysed by Western blotting. Bands corresponding to Mas1 and Mas2 were detected in the whole cell extracts, crude and pure mitochondrial extracts (Figure 5B). The inner mitochondrial protein, Atp2, served as a positive control, being detected in the same samples (Figure 5B). The purity of the mitochondrial fractions was determined by probing for the cytosolic protein Hog1, which was abundant in the whole cell extracts and post-mitochondrial (PMF) fractions, but is extremely scarce in the enriched mitochondrial fractions (Figure 5B). Lastly, we performed an assay to determine where Mas1 is localised in the mitochondria. The organelles were purified from the cells expressing Mas1-Myc and subjected to a series of treatments with or without addition of the exogenous proteinase K. Similar to Aac2, an inner mitochondrial membrane protein, Mas1-Myc remained inaccessible to proteinase K (PK) both in intact and osmotically disrupted (Swelling) organelles. However, both proteins were degraded when osmotically challenged mitoplasts were incubated with PK in the presence of dodecyl maltoside (Detergent), which disrupts the inner mitochondrial membrane. These data strongly suggest that Mas1 is a mitochondrial matrix protein (Figure 5C). Similar results were obtained for the C-terminally Myc-tagged Mas2 (data not shown).
–
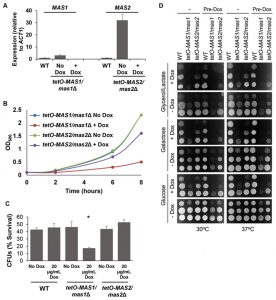
As Mas1 and Mas2 are essential proteins in S. cerevisiae [46], we generated conditional mutants to assess mitochondrial morphology and function. First, MAS1 and MAS2 were placed under the tetO promoter and gene expression was assessed. Growth in the presence of doxycycline significantly reduced expression of MAS1 and MAS2 in the tetO-MAS1/mas1Δ and tetO-MAS2/mas2Δ strains compared to wild-type cells (Figure 6A). Second, viability was assessed, whereby we observed that both strains exhibited reduced growth (Figure 6B), but remained viable during phenotypic analysis. Therefore, the tetO promoter sufficiently reduced expression of MAS1 and MAS2, however, it has been previously reported that doxycycline affects mitochondrial function [47]. As such, to ensure that results were not affected due to the addition of doxycycline, all experiments also included a doxycycline only control.
–
Defects in mitochondrial function cause hypersensitivity to oxidative stress and impaired capacity to grow on non-fermentable carbon sources in S. cerevisiae [48]. To determine if depletion of MAS1 or MAS2 also confers hypersensitivity to oxidative stress, as was the case with deletion of YDJ1 (Figure 1C), we monitored survival of our tetO-MAS1/mas1Δ and tetO-MAS2/mas2Δ strains in response to oxidative stress induced by treatment with 5 mM H2O2 for one hour after depletion of target gene expression. We observed a significant decrease (p<0.05) in cell survival in response to oxidative stress upon depletion of MAS1, but not MAS2 (Figure 6C), likely due to a more auxiliary role of the latter protein. Growth of our tetO-MAS1/mas1Δ and tetO-MAS2/mas2Δ strains was also measured on fermentable medium, YPD (contains 2% glucose as a sole carbon source), fermentable medium that requires mitochondrial function, YP-galactose (2%), and non-fermentable YP-Glycerol/Lactate (each at 2%). Strains were grown in YPD or YPD containing 20 μg/ml doxycycline for 24 hours before being spotted on YP-based plates with or without 20 μg/ml doxycycline containing glucose, galactose or glycerol/lactate (Figure 6D). Plates were incubated at 30°C or 37°C. The wild-type strain grew well on all carbon sources (Figure 6D). However, both the tetO-MAS1/mas1Δ and tetO-MAS2/mas2Δ strains exhibited growth defects on YP-galactose and YP-glycerol/lactate in the presence of doxycycline (Figure 6D). Strikingly, the tetO-MAS1/mas1Δ strain pre-grown in doxycycline and plated on YP-galactose or YP-glycerol/lactate in the presence of doxycycline failed to grow (Figure 6D). Thus, our observations suggest that depletion of MAS1 or MAS2 perturbs mitochondrial function in C. albicans.
–
Ydj1 is important for mitochondrial import and function
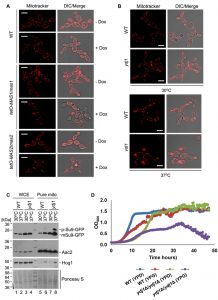
To determine if Ydj1 plays any role in mitochondrial function in C. albicans, we first examined mitochondrial morphology. First, we grew wild-type, tetO-MAS1/mas1Δ and tetO-MAS2/mas2Δ strains in YPD in the presence or absence of doxycycline for 24 hours and then subcultured for an additional 4 hours under the same conditions with the addition of mitochondria-specific vital fluorescent dye Mito Tracker Red. Generally, mitochondria form dynamic tubular networks capable of changing shape and moving throughout the cell. Doxycycline-mediated transcriptional repression of MAS1 or MAS2 induced punctate, aggregated mitochondrial structures, reflecting dysfunction-induced fragmentation of the mitochondrial network (Figure 7A). Second, we looked at the ydj1Δ/ydj1Δ mutant strains in YPD with MitoTracker Red. At 30°C, the ydj1Δ/ydj1Δ mutant presented similarly to the wild type, with only a few punctate, aggregated mitochondrial structures (Figure 7B). Given that the severity of mitochondrial import phenotypes can be exacerbated at elevated temperature [14], we monitored mitochondrial network morphology at 37°C. At 37°C, we observed significant differences in the network morphology in the ydj1Δ/ydj1Δ mutant compared to wild-type cells (Figure 7B). The ydj1Δ/ydj1Δ mutant cells no longer contained tubular mitochondrial networks, but instead mitochondria were punctate, and often aggregated at the septum of the cell (Figure 7B). Thus, Ydj1 is required for normal mitochondrial morphology at elevated temperatures.
–
Next, to more explicitly test whether Ydj1 influences the import of mitochondrial proteins, we used the reporter construct comprising targeting signal (amino acid residues 1-69) of the ATP synthase subunit 9 (Su9) from Neurospora crassa and the GFP moiety [49]. Su9(1-69) is a well characterised mitochondrial matrix targeting sequence which includes a processing site for the MPP Mas1 and Mas2, and three amino acids of the mature form of Su9 [50]. We hypothesised that in a strain lacking Ydj1, the precursor form of Su9-GFP (predicted to be 37 kDa), would accumulate, as Ydj1 is required to deliver proteins to Mas1 and Mas2 for cleavage. However, in wild-type cells, the precursor form would be cleaved upon import into the mitochondria, leading to the mature form (predicted to be 29 kDa). Wild-type and ydj1Δ/ydj1Δ mutant strains with the Su9-GFP construct were grown at 30°C or 37°C, and proteins were extracted from whole cells and pure mitochondria. Upon probing the whole cell extract for Su9-GFP, only the mature form was detected in both the wild type and ydj1Δ/ydj1Δ mutant (Figure 7C). However, the pure mitochondrial extracts revealed an accumulation of the precursor form of Su9-GFP in the ydj1Δ/ydj1Δ mutant at 37°C compared to wild type (Figure 7C). This is likely due to the fact that mitochondrial import phenotypes can be exacerbated at elevated temperature due to lower efficiency of substrate translocation [14]. Thus, Ydj1 is required for full maturation of Su9-GFP, likely through its association with the MPP Mas1 and Mas2.
–
The data thus far suggests that Ydj1 plays a role in mitochondria import. As such, we hypothesised that mitochondrial function would be impaired in the ydj1Δ/ydj1Δ mutant. To test this, we assessed the growth of the wild-type and ydj1Δ/ydj1Δ mutant strains in YP-glucose (2%) and YP-galactose (2%), finding that the ydj1Δ/ydj1Δ mutant was unable to maintain sustained growth in YP-galactose (Figure 7D). Thus, Ydj1 is required for normal mitochondrial morphology, respiration and import of mitochondrial proteins.
–
Ydj1 is localised to the cytosol and outer mitochondrial membrane
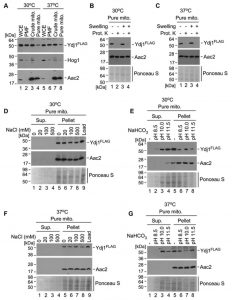
In S. cerevisiae, the majority of Ydj1 is found in the cytoplasm and endoplasmic reticulum, with a small fraction localising to mitochondria [13]. Given the interaction of Ydj1 with the MPPs, which are mitochondrial proteins, and the phenotypes we have observed with defective mitochondrial morphology, import and growth on non-fermentable carbon sources, we proposed that Ydj1 does, in part, localise to the mitochondria in C. albicans. To assess this, we probed for Ydj1 from whole cell extracts, post-mitochondrial fractions, crude mitochondria, and pure mitochondria isolated from the FLAG tagged Ydj1 strain grown at 30°C and 37°C. Ydj1 was present in all fractions (Figure 8A), suggesting that it is present in the cytosol and mitochondria. However, the levels of Ydj1 in the pure mitochondrial fraction were lower than the levels of Ydj1 seen in all other subcellular fractions.
–
To determine the mitochondrial compartment to which Ydj1 localises, gradient purified mitochondria were mock treated or subjected to osmotic disruption (Swelling) of the outer membrane, followed by addition of the exogenous Proteinase K. Addition of Proteinase K causes a loss of Ydj1 in swollen and mock-treated mitochondria isolated at both 30°C (Figure 8B) and 37°C (Figure 8C), suggesting an association with the cytosolic side of the outer membrane. To assess this association further, we performed subcellular fractionations of Ydj1-containing mitochondria under salt and alkaline conditions. Western blotting analysis of salt extracts demonstrates the presence of Ydj1 and the control inner membrane-anchored protein Aac2 only in pelleted fractions isolated from gradient-purified mitochondria from the cells grown at both 30°C and 37°C (Figure 8D and 8F), indicating that Ydj1 is firmly bound to the outer mitochondrial membrane. Conversely, immunoblotting analysis of alkaline extracts revealed the majority of Ydj1 signal – but not Aac2 signal – in the supernatant fractions (Figure 8E and 8G), reflecting the peripheral association of Ydj1 with mitochondria. Taken together, these data suggest that a pool of Ydj1 is specifically and tightly associated with the cytosolic side of the outer mitochondrial membrane.
DISCUSSION
As mitochondria evolved, the mitochondrial genome was reduced and has almost completely been incorporated into the nuclear genome. As such, most mitochondrial proteins are synthesised in the cytosol with a cleavable N-terminal sequence that targets them for import with the assistance of molecular chaperones. To date, the identity and function of mitochondrial import proteins in C. albicans has remained largely unknown. We identified a novel role for the C. albicans Hsp40 chaperone Ydj1 in mitochondrial import, implicating the mitochondrial peptidases Mas1 and Mas2 as central to this process.
–
Chaperones are fundamental to orchestrating protein trafficking and maintaining protein homeostasis during stress in the eukaryotic kingdom [51]. For example, Hsp90 promotes the folding and function of key substrate proteins [24][52][53], often in collaboration with co-chaperones [54]. Hsp90 has been well studied in C. albicans [22][24][26][55], but the roles of other chaperones remain largely unknown. For example, the Hsp40 chaperone Ydj1, the most studied Hsp40 chaperone in S. cerevisiae, had not been characterised in C. albicans. Utilising genetic and biochemical approaches, we have uncovered a plethora of diverse phenotypes related to stress responses upon loss of Ydj1 (Figure 1). These phenotypes may be attributable to the role of Ydj1 in promoting protein homeostasis. Consistent with this, our proteomic analysis revealed that Ydj1 interacts with eight other chaperones or co-chaperones, some of which have been previously characterised in S. cerevisiae, corroborating our findings. Indeed, a strong interaction was observed between Ydj1, Hsp104, Ssa2 and Hsp70, similar to findings in S. cerevisiae, where Ydj1 interacts with Hsp104 and Ssa1, together aiding in protein re-folding [41]. Hsp104 and Hsp70 are both required for high temperature growth and virulence in C. albicans [56][57][58]. Thus, the increase in stress sensitivity in the C. albicans ydj1Δ/ydj1Δ mutant could be due to an accumulation of misfolded proteins and a deficiency in re-folding aggregated proteins that occur during stress.
–
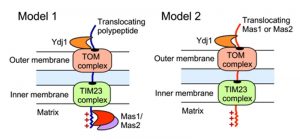
The impact of Ydj1 on cellular stress responses may also be mediated through control of mitochondrial function, which is crucial for cellular stress survival [27], and is contingent upon the proper import of proteins with a targeting signal sequence. We detected a strong interaction between Ydj1 and the MPP Mas1 and Mas2, both in the presence and absence of heat shock (Figure 3). In S. cerevisiae, Ydj1 functions together with Ssa1 to mediate the import of mitochondrial preproteins [14], but the precise mechanisms of targeting and import of mitochondrial proteins remains elusive. Mitochondria contain transport machineries in both the outer (TOM) and inner (TIM) membranes to import nuclear-encoded proteins, whereby they are cleaved by MPP [59]. We established that Ydj1 is tightly associated with the mitochondrial outer membrane (Figure 8), and that Ydj1 from gradient purified mitochondria physically interacts with the MPP Mas1 and Mas2 (Figure 5A). Furthermore, ydj1Δ/ydj1Δ mutant cells displayed defective mitochondrial morphology and were unable to sustain growth on media containing a non-fermentable carbon source, suggesting that Ydj1 plays a role in mitochondrial function (Figure 7B and 7D). Utilising the model mitochondrial import protein, Su9-GFP, which is processed by Mas1 and Mas2 in the mitochondria, we found that deletion of YDJ1 leads to an accumulation of the immature form of Su9-GFP at 37°C (Figure 7C). Thus, Ydj1 may interact with the MPPs indirectly, via binding with the MPPs substrate proteins during their import, or it may be a direct interaction, binding the MPP subunits upon their own translocation to the mitochondria (Figure 9). Further analysis with other MPP substrate proteins would be required to resolve the nature of the interaction.
–
We found that Ydj1 also interacts with other key players involved in mitochondrial import and function, including Hsp70, Mdj1 and Hsp78. Cytosolic Hsp70 proteins were the first chaperones implicated in mitochondrial protein import [60], with studies in yeast utilising synthetic peptides revealing that presequence peptides can bind cytosolic Hsp70 [61]. The mitochondrial DnaJ (Mdj1) is a soluble mitochondrial matrix protein that binds precursor proteins entering the matrix in the latter stages of import, aiding in their folding [62]. Similar to Mdj1 is the soluble mitochondrial matrix protein Hsp78, which binds misfolded polypeptides in the matrix to prevent aggregation [63]. We observed an interaction between Ydj1 and Hsp78 during heat shock, which impairs mitochondrial protein synthesis [63], suggesting that Ydj1, together with Hsp78 promotes re-folding of damaged proteins upon heat shock. Each of these chaperones are regulated by the heat shock transcription factor Hsf1 in C. albicans [26]. In S. cerevisiae, Hsf1 is required for import of mitochondrial proteins, likely attributable to regulatory control of Ydj1 and other chaperones [7][8]. Thus, Ydj1 may promote survival in response to stress through interactions with co-chaperones, thereby promoting protein re-folding and mitochondrial function, which are both required for survival in response to cellular stresses [27].
–
As a consequence of their biochemical function in modulating protein homeostasis, molecular chaperones also have central roles in orchestrating temperature-dependent developmental programs. Temperature regulates numerous cellular processes in C. albicans, including a morphological transition from the yeast form at ambient temperatures to filamentous forms in response to thermal stress; this transition is important for dissemination, tissue penetration, immune evasion, and virulence [18]. The expression of many molecular chaperones increases in response to elevated temperature [64], and the molecular chaperone Hsp21 serves as a regulator of morphogenesis [38]. Thus, we hypothesised that Ydj1 might interact with and promote folding of a positive regulator of filamentous growth. Although none of the interactors we identified were strictly required for morphogenesis, as was Ydj1, we detected an interaction with three proteins that promote robust filamentation; the small heat shock protein Hsp21 [38], as well as Cdc48 and Mas1, proteins both involved in mitochondrial function [5][45] (Figure 4). Notably, the interaction of Ydj1 with Hsp21 and Cdc48 increased with heat shock, suggesting that the regulation of temperature-dependent filamentation may occur in part through these regulators. Given that inhibition of mitochondrial function blocks C. albicans morphogenesis [32], and Mas1 and Cdc48 are both important for mitochondrial function, an alternative model is that the morphogenetic defect observed in the absence of Ydj1 may be attributable to reduced mitochondrial protein import. Thus, as a central regulator of mitochondrial function, cellular stress responses, and virulence traits in a leading fungal pathogen, Ydj1 provides a novel prospective target for antifungal drug development.
MATERIALS AND METHODS
Strains and growth conditions
All strains are listed in Table 1. Strains were grown in YPD (1% yeast extract, 2% bactopeptone, 2% glucose) or YPGD (1% yeast extract, 2% bactopeptone, 0.2% glucose, 2% galactose) [65] with shaking at 200 rpm. Doxycycline (BD Biosciences) was added to YPD medium at a concentration of 0.05 µg/ml or 20 µg/ml, as stated. MitoTracker Red (Life Technologies, M-7512) was added at 100 nM. To repress expression of MAS1 or MAS2 from the tetracycline-repressible (tetO) promoter, overnight cultures in YPD at 30°C were diluted to OD600 0.2 in the absence or presence of doxycycline and cultured for 24 hours. Cells were diluted to OD600 0.05 in the same conditions and grown to mid-log phase (~5 hours) prior to microscopy, RNA extraction, or Western blot analysis. A 30°C – 42°C heat shock was imposed, as indicated, following our established protocols [21].
 |
–
Strain construction
To generate an YDJ1 homozygous deletion mutant, the NAT flipper cassette (pLC49, Table 2) [66] was PCR amplified with oLC3129/oLC3131 containing sequence homologous to the upstream and downstream regions of YDJ1. The PCR product was transformed into the wild-type strain SN95 (CaLC239), and NAT-resistant transformants were PCR tested with oLC275/oLC3132 and oLC274/oLC3133 to verify integration of the cassette. The NAT cassette was excised, generating CaLC3124 (YDJ1/ydj1Δ). A second round of PCR was performed to delete the second allele by re-amplifying the NAT flipper cassette with oLC3128/oLC3130. The PCR product was transformed into CaLC3124 and NAT-resistant transformants were PCR tested with oLC275/oLC3132 and oLC274/oLC3133 to verify integration of the cassette, and oLC3134/oLC3135 to verify loss of the wild-type allele, creating CaLC3160 (ydj1Δ/ydj1Δ).
 |
–
To regulate MAS1 and MAS2, one allele of MAS1 or MAS2 was deleted, and the other was placed under the control of the tetO promoter that can be repressed with tetracycline or the analog doxycycline. Briefly, the NAT flipper cassette (pLC49), containing sequence homologous to the upstream and downstream regions of MAS1 or MAS2 was PCR amplified with oLC4284/oLC4285 (MAS1) or oLC4295/4296 (MAS2). The PCR product was transformed into the wild-type strain SN95 (CaLC239), and NAT-resistant transformants were PCR tested with oLC275/oLC4286 and oLC274/oLC4283 (MAS1) or oLC275/oLC4297 and oLC274/oLC4294 (MAS2) to verify integration of the cassette. The NAT cassette was then excised to create CaLC4390 (MAS1/mas1D) or CaLC4391 (MAS2/mas2D). The tetracycline-repressible transactivator, the tetO promoter, and the NAT flipper cassette were PCR amplified from pLC605 (Table 2) [67] using oligos oLC4284/oLC4287 (MAS1) or oLC4295/oLC4298 (MAS2). The PCR product was transformed into CaLC4390 (MAS1/mas1D) or CaLC4391 (MAS2/mas2D). Correct integration at the MAS1 locus was verified by colony PCR using primer pairs oLC534/oLC4286 and oLC4288/oLC300, respectively, generating CaLC4410 (tetO-MAS1/mas1Δ). Loss of the wild-type allele was verified with oLC4286/oLC4288. Correct integration for MAS2 was verified using oligos oLC534/oLC4297 and oLC4299/oLC300. Loss of the wild-type allele was confirmed using primers oLC4297/4299, generating CaLC4438 (tetO-MAS2/mas2Δ).
–
To detect Ydj1 interactors by mass spectrometry, Ydj1 was tagged with 2xFLAG at its N-terminus. Our previous attempts to C-terminally tag Ydj1 resulted in a non-functional protein. The YDJ1-ARG4 cassette was PCR amplified from CaLC3924 with long primers containing 2xFLAG (oLC3940/3807). The subsequent product was PCR amplified using primers oLC3941/3807, which contain homology upstream and downstream of the YDJ1 locus, and transformed into the ydj1Δ/ydj1Δ strain (CaLC3160). Arginine prototrophic transformants were PCR tested for upstream integration using oLC3132/3828 and downstream integration using oLC3938/1594, generating CaLC3958 (FLAG-YDJ1/ydj1Δ). This strain also acted as a complemented strain for the ydj1Δ/ydj1Δ mutant.
–
To detect Mas1 and Mas2, the proteins were Myc tagged at their C-termini using a Myc-NAT cassette. The Myc-NAT cassette was generated using fusion PCR. Myc-tag with homology to MAS1 or MAS2 and NAT was PCR amplified from pLC578 (Table 2) with oLC5140 and oLC5157 (905 bp) for MAS1 and oLC5142 and oLC5157 (905 bp) for MAS2. The NAT marker with homology to Myc and MAS1 was PCR amplified from pLC49 with oLC5156 and oLC5158 (977 bp). The NAT marker with homology to Myc and MAS2 was PCR amplified from pLC49 with oLC5156 and oLC5159 (977 bp). The PCR product was purified by spin column purification and 1:100 dilution was used as templates for the fusion PCR at a 1:1 molar ratio (~0.4 ng of the longer template). For Mas1, the Myc-NAT cassette was amplified with oLC5140 and oLC5158 and transformed into CaLC3958. Correct integration at the C-terminus of MAS1 was verified by amplifying across both junctions using primer pairs oLC4282 + oLC2029 (upstream 632 bp) and oLC4283 + oLC3849 (downstream 955 bp), generating CaLC4967. For Mas2, the Myc-NAT cassette was amplified with oLC5142 and oLC5159 and transformed into CaLC3958. Correct integration at the C-terminus of MAS2 was verified by amplifying across both junctions using primer pairs oLC4293 + oLC2029 (upstream 783 bp) and oLC4294 + oLC3849 (downstream 1106 bp), generating CaLC4969.
–
The ACT1p-SnSu9-GFP-NAT cassette was generated using fusion PCR. ACT1p-SnSu9 was PCR amplified from the Su9-GFP plasmid, pYX142 [49], using primers oLC4348/oLC4349. The GFP-NAT cassette was PCR amplified from pLC389 (Table 2) using primers oLC4350/oLC4275. The PCR products were spin column purified and the concentration was measured by nanodrop. The products were diluted 1:20 and added to the PCR mastermix in 1:1 molar ratio (~15 ng of the longer PCR product). The fusion was PCR amplified with oLC4348/oLC4275 and transformed into CaLC239 (SN95), generating CaLC4422 (ACT1p-Su9-GFP-NAT/ACT1). Correct integration at the ACT1 locus was verified by PCR using primer pairs oLC3833/oLC600 and oLC274/oLC3024.
–
The tetO-SnSu9-GFP-NAT cassette was made by PCR amplifying the genomic DNA of CaLC4422 using oLC4384/oLC2644. The PCR product was transformed into CaLC4547 (tetO-CAS5/CAS5) to target integration at the CAS5 locus, generating CaLC4562 (tetOp-Su9-GFP-NAT/CAS5). Correct integration at the tetO-CAS5 locus was verified by PCR amplifying across both junctions using primer pairs oLC2034/oLC534, oLC300/oLC600 and oLC274/oLC2164. CaLC4547 was generated by PCR amplifying the tetracycline-repressible transactivator, tetO promoter and NAT flipper cassette from pLC605 using primers oLC2088/oLC2089 and transformed into CaLC239. Correct upstream and downstream integration at the CAS5 locus was verified by PCR amplifying across both junctions using primer pairs oLC2034/oLC534 and oLC300/oLC2145.
–
To express Su9 in the ydj1Δ/ydj1Δ mutant, first the tetracycline-repressible transactivator and tetO promoter were PCR amplified from pLC605 using primers oLC2088/oLC2089 and transformed into CaLC3160 (ydj1Δ/ydj1Δ), generating CaLC4554 (ydj1Δ/ydj1Δ tetO-CAS5/CAS5). Correct upstream and downstream integration at the CAS5 locus was verified by PCR using primer pairs oLC2034/oLC534 and oLC300/oLC2145. Second, the tetO-SnSu9-GFP-NAT cassette was PCR amplified from the genomic DNA of CaLC4422 using oLC4384/oLC2644 and transformed into CaLC4554. Correct integration at the tetO-CAS5 locus was verified by PCR amplifying across both junctions using primer pairs oLC2034/oLC534 and oLC274/oLC2164. All primers are listed in Table 3.
 | TABLE 3. Primers. |
–
Phenotypic assays
The susceptibilities of strains to stress were determined with a constant concentration of the following stressors: calcofluor white (CFW: 25 µg/ml), NaCl (1 M), H2O2 (5 mM), and temperature (30°C, 37°C, or 42°C). All stresses were assayed in flat-bottom 96-well microtiter plates (Starstedt) in a final volume of 200 µl/well. Overnight cultures were diluted in YPD to ~103 cells/ml and used to inoculate the first well. Cells were subsequently diluted ten-fold in YPD in wells across the plate. Plates were wrapped in tin foil and incubated statically at 30°C (except for temperature screens) for 48 or 72 hours. Cells were re-suspended and final cell densities were determined by measuring OD600 using a spectrophotometer. Data was quantitatively plotted with colour using Java Treeview 1.1.3 (http:// jtreeview.sourceforge.net). Assays were performed in duplicate on three different occasions.
–
Minimum inhibitory concentration assay
Antifungal tolerance was determined in flat-bottom, 96-well microtiter plates (Starstedt). MICs were set up to a final volume of 200 µl/well with two-fold dilutions of caspofungin (Merck) and the last well containing no caspofungin. Overnight cultures were diluted in YPD such that ~103 cells were inoculated into each well. Plates were wrapped in tin foil and incubated at 30°C for 48 hours. Cells were re-suspended and final cell densities were determined by measuring OD600 using a spectrophotometer. Caspofungin tolerance was tested in duplicate on three different occasions. Data was quantitatively plotted with colour using Java Treeview 1.1.3.
–
Microscopy
Filamentation was observed by subculturing overnight cultures of indicated strains into YPD medium in the absence or presence of 10% newborn bovine serum (Gibco #26010-066) at an OD600 0.1 and growing cells at 30°C or 37°C for four hours. For high temperature filamentation, overnight cultures were subcultured into YPD at an OD600 0.1 and grown with shaking at 30°C or 39°C for three hours. Imaging was performed on a Zeiss Imager M1 upright microscope and AxioCam MRm with AxioVision 4.7 software.
–
Mitochondrial morphology in the wild-type, ydj1Δ/ydj1Δ, tetO-MAS1/mas1Δ and tetO-MAS2/mas2Δ strains was assessed by subculturing an overnight culture of each strain into YPD at an OD600 0.1 in the absence or presence of 20 μg/ml doxycycline for 24 hours. Strains were subcultured again in the same conditions with the addition of 100 nM MitoTracker and grown for a further four hours at 30°C or 37°C before imaging. Mitotracker Red-stained live cells were visualized as described before [29] using the Olympus IX81-FV5000 confocal laser scanning microscope at 543-nm laser line with a 100× oil lens. Images were acquired and processed with Fluoview 500 software (Olympus America).
–
qRT-PCR
To ensure depletion of MAS1 and MAS2 transcript levels, strains SN95 (CaLC2993), CaLC4410 (tetO–MAS1/mas1∆) and CaLC4438 (tetO-MAS2/mas2∆) were grown overnight at 30°C in YPD with shaking at 200 rpm. Stationary phase cultures were split, adjusted to an OD600 of 0.1 where one culture was treated with 20 μg/ml doxycycline, while the other remained untreated. Cells were grown for 24 hours at 30°C. After 24 hours, cultures were re-inoculated into YPD +/- doxycycline as per the previous growth condition. Cells were grown for 5 hours at 30°C and 10 ml was harvested from each culture, centrifuged at 3000 rpm for 2 minutes at 4°C, washed once with dH2O before being frozen at -80°C. RNA was subsequently isolated using the QIAGEN RNeasy kit and cDNA synthesis was performed using the AffinityScript cDNA synthesis kit (Stratagene). qRT-PCR was carried out using the Fast SYBR Green Master Mix (Thermo Fisher Scientific) in 384-wells with the following cycle conditions: 95°C for 3 minutes, 95°C for 10 seconds and 60°C for 30 seconds for 39 rounds, 95°C for 10 seconds, 65°C for 5 seconds. All reactions were performed in triplicate using the following primer pairs: MAS1 (oLC4377/4288), MAS2 (oLC4376/4299). Transcript levels were normalised to ACT1 (oLC2285/2286). Data were analysed using the BioRad CFX Manager software, version 3.1 (BioRad).
–
Growth assays
For the spot assays, wild-type (SN95), tetO-MAS1/mas1Δ and tetO-MAS2/mas2Δ strains were grown in the absence or presence of 20 μg/ml doxycycline in YPD for 24 hours. The cells were then serially diluted (with the lowest dilution being OD600 1) and plated on YP-based plates containing glucose (2%), galactose (2%) or glycerol/lactate (each at 2%), with or without doxycycline. Plates were incubated at 30°C or 37°C for 36 hours and imaged. For the growth curves, overnight cultures of wild type or ydj1Δ/ydj1Δ grown in YPD at 30°C were subcultured into YPGD at an OD600 of 0.1 for 24 hours at 30°C. Growth kinetics were measured by inoculating cells from the cultures grown in YPGD at 30°C to an OD600 of 0.0625 in 100 µl of YP with 2% glucose or 2% galactose in flat bottom, 96-well microtiter plates (Sarstedt). Cells were grown in a Tecan GENios microplate reader (Tecan Systems Inc.) at 30°C with orbital shaking. Optical density measurements were taken every 15 minutes for 48 hours. Statistical significance was evaluated using GraphPad Prism 6.01.
–
Western blotting
Proteins were extracted and subjected to Western blotting by pelleting cells at an OD600 0.8 by centrifugation, washing with sterile H2O, and resuspending in 50 μl of 2x sample buffer (0.35 M Tris-HCl, 10% (w/w) SDS, 36% glycerol, 5% β-mercaptoethanol, and 0.012% bromophenol blue). Extracts were loaded in wells of a 6% or 10% SDS–PAGE gel. Separated proteins were transferred to a PVDF membrane for 1 hour at 100 V at 4°C. Membranes were blocked in 5% milk in PBS containing 0.1% Tween-20 at room temperature for 1 hour and subsequently incubated in primary antibody for one hour at room temperature in PBS-T+5% milk. Membranes were washed with 1x PBS-T and probed for one hour with secondary antibody dissolved in 1x PBS-T+5% milk (PBS, 0.1% Tween-20, 5% (w/v) milk). Membranes were washed in PBS-T and signals detected using an ECL Western blotting kit as per the manufacturer’s instructions (Pierce).
–
FLAG-tagged Ydj1 was detected using a 1:10,000 dilution of anti-FLAG HRP conjugated antibody (Sigma, A8592). To detect GFP a 1:1,000 dilution of anti-GFP (Santa-Cruz, sc-8334) was used. To detect Tubulin, an anti-Tub1 antibody was used (AbD Serotec, MCA78G) at a 1:5,000 dilution. To detect Myc, an anti-Myc (mouse) (11667149001, Roche Diagnostics) at 1:1,000 or anti-Myc (rabbit) (A-14, Santa Cruz Biotechnology) at 1:2,500 was used. To detect Aac2, an anti-Aac2 (rabbit) was used at a 1:5,000 dilution, a gift from Dr. Carla Koehler (UCLA). To detect Atp2, an anti-Atp2 (rabbit) serum was used at a 1:1,000 dilution, a gift from Dr. Alex Tzagoloff (Columbia University). Hog1 was detected using anti-Hog1 (y215, Santa Cruz Biotechnology) at 1:2,500. Secondary antibodies were anti-rabbit (7074S, Cell Signaling) at 1:5000, and anti-mouse (115-035-068, Jackson ImmunoResearch Laboratories, Inc.) at 1:5000.
–
Ydj1 affinity purification
Untagged wild-type (SN95) and 2xFLAG-YDJ1/ydj1Δ (CaLC3958) cells were grown overnight at 30°C in YPD. Stationary phase cultures were split, adjusted to an OD600 of 0.1 in YPD and grown to an OD600 of 0.8. Cultures were split and subjected to a 15 minute 30°C – 42°C heat shock, exactly as previously described [21]. Cells were harvested at 4000 rpm for 10 minutes at 4°C, washed twice with ice-cold 1x PBS and snap frozen in liquid nitrogen. A second biological replicate was performed on a separate date. One-step affinity purification of 2xFLAG-Ydj1 was performed using anti-FLAG M2 Magnetic Beads (Sigma-Aldrich), as previously described [68] with some modifications. Briefly, Pellets from 250 ml of cultured C. albicans yeast cells were resuspended in 10 ml of lysis buffer (100 mM HEPES, pH 8.0, 20 mM magnesium acetate, 10% glycerol (v/v), 10 mM EGTA, 0.1 mM EDTA, 0.4% NP-40 supplemented with fresh protease inhibitors mixture (100 fold dilution; P8215 (Sigma-Aldrich) and 1 mM PMSF), and subsequently lysed by bead-beating for 3 x 2 minutes in a cold room. The lysates were then sonicated for 3 x 30 second pulses at 65% amplitude using a QSONICA 125W sonicator equipped with 1/8” probe. The lysates were then centrifuged at 1,800 xg for 10 minutes and the resulting supernatant transferred to a fresh tube. Freshly washed anti-FLAG M2 magnetic beads (30 μl slurry per sample) were added and the samples were incubated with end-over-end rotation for 3 hours at 4°C. Using a DynaMag-2 magnet (ThermoFisher), the beads were collected on the side of the sample tube and the supernatant was discarded. The beads were washed three times by resuspension in 1 ml of wash buffer (100 mM HEPES, pH 7.4, 20 mM magnesium acetate, 10% glycerol (v/v), 10 mM EGTA, 0.1 mM EDTA, 0.5% Nonidet P-40) and transferred to a fresh 1.5 ml tube during the last wash. Finally, the beads were washed with 1 ml of 20 mM Tris-HCl pH8, 2 mM CaCl2. Following the last wash, the samples were quickly centrifuged and the last drops of liquid removed with a fine pipette. The now-dried beads removed from the magnet were resuspended in 7.5 µL of 20 mM Tris-HCl (pH 8.0) containing 750 ng of trypsin (Sigma-Aldrich; Trypsin Singles, T7575), and the mixture was incubated at 37°C with agitation overnight (~15 hours). After this first incubation, the sample was magnetized and the supernatant transferred to a fresh tube. Another 250 ng of trypsin was added (in 2.5 µL of 20 mM Tris-HCl (pH8)), and the resulting sample was incubated at 37°C for 3 – 4 hours without agitation. Formic acid was then added to the sample to a final concentration of 2% (from a 50% stock solution) and the samples were stored at -40°C until analyzed by mass spectrometry. Pellets prepared from untagged parental strains were used as negative controls and processed in parallel to Ydj1 for the AP-MS experiments.
–
Mass spectrometry and data analysis
Ydj1 AP-MS samples and controls were analyzed by mass spectrometry in two biological replicates. 5 µl of each sample representing approximately 50% of the samples were directly loaded at 400 nl/min onto a 75 μm x 12 cm emitter packed with 3 µm ReproSil-Pur C18-AQ (Dr. Maisch HPLC GmbH.). Peptides were eluted from the column over a 90-minute gradient generated by a NanoLC-Ultra 1D plus (Eksigent) nano-pump and analyzed on a TripleTOFTM 5600 instrument (AB SCIEX). The gradient was delivered at 200 nl/minute, starting at 2% acetonitrile (all solvents also contain 0.1% formic acid) and ending at 35% acetonitrile over 90 minutes, followed by a 15-minute clean-up in 80% acetonitrile, and a 15-minute equilibration period back to 2% acetonitrile for a total of 120 minutes. To minimize carryover between each sample, the analytical column was washed for 3 hours by running an alternating “saw-tooth” gradient from 35% acetonitrile to 80% acetonitrile, holding each gradient concentration for 5 minutes. Analytical column and instrument performance were verified after each sample by loading 30 fmol BSA tryptic peptide standard (Michrom Bioresources Inc.) with 60 fmol α-Casein tryptic digest and running a short 30 min gradient. TOF MS calibration was performed on BSA reference ions before running the next sample in order to adjust for mass drift and verify peak intensity. The instrument method was set to a Data Dependent Acquisition mode which consisted of one 250 ms MS1 TOF survey scan from 400–1300 Da followed by twenty 100 ms MS2 candidate ion scans from 100 – 2000 Da in high sensitivity mode. Only ions with a charge of 2+ to 4+ that exceeded a threshold of 200 cps were selected for MS2, and former precursors were excluded for 10 seconds after 1 occurrence.
–
Mass spectrometry data generated were stored, searched and analyzed using the ProHits laboratory information management system (LIMS) platform [69]. Within ProHits, the initial WIFF files were first converted to an MGF format using WIFF2MGF converter and to an mzML format using ProteoWizard (v3.0.4468) and the AB SCIEX MS Data Converter (V1.3 beta) and then searched using Mascot (v2.3.02) and Comet (v2012.02 rev.0), respectively. The spectra were searched with the RefSeq database (version 68, November 21th, 2014) acquired from NCBI against a total of 29,524 C. albicans sequences supplemented with “common contaminants” from the Max Planck Institute (http://141.61.102.106:8080/share.cgi?ssid=0f2gfuB) and the Global Proteome Machine (GPM; http://www.thegpm.org/crap/index.html). The database parameters were set to search for tryptic cleavages, allowing up to 2 missed cleavage sites per peptide with a mass tolerance of 40 ppm for precursors with charges of 2+ to 4+ and a tolerance of +/- 0.15 amu for fragment ions. Variable modifications were deamidated asparagine and glutamine and oxidized methionine. The results from each search engine were analyzed through the Trans-Proteomic Pipeline (TPP v4.6 OCCUPY rev 3) [70] via the iProphet pipeline [71]. SAINTexpress (v3.3) [72] was used as a statistical tool to calculate the probability value of each potential protein-protein interaction from background contaminants using default parameters and a ProteinProphet cutoff of 0.95. Controls were kept uncompressed and a FDR of 1% or lower was required for proteins to be classified as significant interaction partners of Ydj1.
–
Dot plots and heat map of Ydj1 interaction networks obtained by AP-MS were generated using a web-based custom-built tool [73]. All RAW mass spectrometry data and downloadable identification and SAINTexpress results tables are deposited in the MassIVE repository housed at the Center for Computational Mass Spectrometry at UCSD (http://proteomics.ucsd.edu/ProteoSAFe/datasets.jsp). The dataset has been assigned the MassIVE ID MSV000079442 and is available for FTP download at: ftp://MSV000079442@massive.ucsd.edu. The dataset will be locked until publication using the password “Candida”. The dataset was assigned the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) identifier PXD003421.
–
Alkaline lysis of yeast cells (whole-cell extracts, WCE)
Yeast whole-cell extracts were prepared according to Khalimonchuk et al. [74]. Briefly, cells corresponding to A600 of 1.0 were pelleted by quick centrifugation (15 sec × 15,000 xg) and resuspended in 250 µl of 50 mM Tris-HCl, pH 8.0. Then, 50 µl of lysis buffer (10 M NaOH, 7.5% β-mercaptoethanol, 2 mM phenylmethylsulfonyl fluoride, PMSF) was added, and samples were vigorously mixed, followed by incubation on ice for 15 min. Then 220 µl of cold 100% TCA was mixed with the lysates, which were incubated on ice for a further 15 min. The samples were centrifuged at 10 min × 20,000 xg at 4°C. The pellets were washed twice with absolute acetone, dried on ice for 20 min, and resuspended in 133 µl of 1× Laemmli buffer followed by incubation at 85°C for 10 min.
–
Isolation and purification of mitochondria
Mitochondria were isolated from 1 L cultures according to the previously described protocol [75] with slight modifications. The 2×FLAG-YDJ1/ydj1Δ yeast strain was pre-grown in 50 ml of YPD for 16 hours. The pre-cultures were then split and used to re-grow in 1 L of YPD at 30°C and 37°C for 6 hours for further isolation and purification of mitochondria. Cells were collected by centrifugation for 5 minutes at 3,500 g at room temperature and then washed once with water. The wet weight of the pellets was then determined. Cells were subsequently resuspended in 0.1 M Tris-SO4, pH 9.4, 10 mM dithiothreitol (DTT) following the ratio of 0.5 g wet weight per 1 ml, and incubated at 30°C for 10 min. The cells were collected by centrifugation and washed once with 20 ml of 1.2 M sorbitol. The pellets were resuspended in 1.2 M Sorbitol, 20 mM KH2PO4, pH 7.4 with the ratio of 0.15 g wet weight per 1 ml. Lyticase (4 mg/g wet weight) was added to the cell suspension, and incubated at 30°C with gentle shaking (80 rpm) for 60 min. Obtained spheroplasts were collected by centrifugation and washed once in 30 ml of 1.2 M sorbitol. The pellets were resuspended in 10 ml of 0.65 M sorbitol, 10 mM Tris-HCl, pH 7.4, 1 mM PMSF and homogenized by 15 strokes in a tight-fitting Dounce homogenizer (40 ml, Kontes Glass Co.) on ice. The homogenates were centrifuged for 5 min at 3,500 xg at 4oC. The supernatants were collected into separate tubes and the pellets were re-homogenized a further two times. Combined supernatants were centrifuged for 10 min at 12,000 xg at 4oC. Supernatants were discarded, and accumulated lipids on the walls of the tubes were removed with paper towels. The remaining pellets were gently resuspended in 10 ml of 0.65 M sorbitol, 10 mM Tris-HCl, pH 7.4, 1 mM PMSF and centrifuged for 5 min at 2,500 xg at 4°C. Obtained supernatants were carefully transferred into new tubes and precipitated by centrifugation 10 min at 12,000g at 4°C. The supernatants were discarded and the pelleted mitochondria were resuspended in 750 ml of 0.65 M sorbitol, 10 mM Tris-HCl, pH 7.4, 1 mM PMSF.
–
Pure mitochondria were obtained by ultracentrifugation through a Histodenz (Sigma-Aldrich) gradient. The gradient was formed by adding 5 ml of 22% (w/v) Histodenz in 1.2M sorbitol and 10 ml of 14% (w/v) Histodenz in 1.2M sorbitol with a load of 1.5 ml crude mitochondria in 0.65 M sorbitol, 10 mM Tris-HCl, pH 7.4, 1 mM PMSF. Samples were centrifuged in a Beckman Coulter OptimaÔ L-100K ultracentrifuge at 50,000 xg for 90 min at 4°C. The fractions of pure mitochondria were then collected from the interphase between 14% and 22% Histodenz, transferred into new tubes and filled up to 30 ml with 0.65 M sorbitol, 10 mM Tris-HCl, pH 7.4, 1 mM PMSF. The samples were precipitated by centrifugation for 10 min at 12,000 xg at 4°C. Obtained pellets were then resuspended in 1.5 ml of 0.65 M sorbitol, 10 mM Tris-HCl, pH 7.4, 1 mM PMSF and precipitated by centrifugation. Pure mitochondria were resuspended in 500 μl of 0.65 M sorbitol, 10 mM Tris-HCl, pH 7.4, 1 mM PMSF. Protein concentrations were measured by Bradford assay.
–
Mitochondrial subfractionation
Mitochondrial protein topology was analyzed as described by Khalimonchuk et al. [76]. Six aliquots of isolated mitochondria (20 µg/sample) were resuspended in 1 ml of 0.65 M Sorbitol, 10 mM Tris-HCl, pH 7.4, and centrifuged for 10 min at 12,000 xg at 4°C. The pellets were resuspended in the following order: 1) intact mitochondria in 500 μl of 0.65 M Sorbitol, 10 mM Tris-HCl, pH 7.4; 2) intact mitochondria in 500 µl of 0.65 M Sorbitol, 10 mM Tris-HCl, pH 7.4 with 5 μl Proteinase K (100 µg/ml); 3) hypotonic swelling to selectively rupture the outer mitochondrial membrane in 500 μl of 20 mM HEPES, pH 7.4; 4) placed in 500 μl of 20 mM HEPES, pH 7.4 with 5 μl Proteinase K (100 μg/ml); 5) lysed mitochondria in 450 μl of 20 mM HEPES, pH 7.4 with 50 μl 10% n-dodecyl-β-D-maltoside (DDM); 6) placed in 450 μl of 20 mM HEPES, pH 7.4 with 50 μl 10% DDM and 5 μl Proteinase K (100 μg/ml). All samples were kept on ice for 30 min with gentle periodical mixing. After incubation 2.5 µl of PMSF (200 mM) was added to all samples. This was then followed by precipitation with 55 μl of 100% (w/v) trichloroacetic acid (TCA) added to every sample and vigorously mixed and treated as described above for whole-cell extract, except for adding DTT to a final concentration of 100 mM to 1× Laemmli buffer.
–
Carbonate extraction was performed as described by Fujiki et al. [77]. Four aliquots of gradient-purified mitochondria (20 μg/sample) were centrifuged for 10 min at 12,000 xg at 4°C. The pellets were resuspended in 500 μl of the following solutions: 1) 0.65 M sorbitol, 10 mM Tris-HCl, pH 7.4, 1 mM PMSF; 2) 0.1 M NaHCO3, pH 8.25, 1 mM PMSF; 3) 0.1 M NaHCO3, pH 10.0, 1 mM PMSF; 4) 0.1 M NaHCO3, pH 11.5, 1 mM PMSF. The samples were incubated on ice for one hour followed by centrifugation at 60,000 xg for 45 min at 4°C. The pellets were resuspended in 40 μl of 1× Laemmli buffer, 100 mM DTT and incubated for 10 min at 85°C. The supernatants were transferred into new tubes and precipitated with 100% TCA as described above.
–
Sodium chloride extraction was performed similarly, however, pellets of four aliquots of gradient-purified mitochondria (20 μg/sample) were resuspended in total volume of 500 μl of 0.65 M sorbitol, 10 mM Tris-HCl, pH 7.4, 1 mM PMSF with the following final concentrations of NaCl: 0, 20, 100, and 500 mM.
–
Postmitochondrial fractions (PMFs) were obtained as following: the amount of cells corresponding to A600 of 2.0 were pelleted by quick centrifugation (15 sec × 15,000 xg) and resuspended in 500 μl of 0.65 M sorbitol, 10 mM Tris-HCl, pH 7.4, 1 mM PMSF. Approximately 300 μl of glass beads (0.5 mm) were added, and the samples were vigorously mixed for 5 min. The supernatants were transferred into new tubes. The beads were washed with 500 μl of 0.65 M sorbitol, 10 mM Tris-HCl, pH 7.4, 1 mM PMSF, and the collected supernatants were combined. The cell extracts were then centrifuged at 2 min x 4,000 xg. Obtained supernatants were carefully transferred into new tubes, and then centrifuged again at 20 min x 20,000 xg. Collected supernatants were then precipitated with TCA as described above.
–
Co-immunoprecipitation assay
Mitochondrial protein interactions were assayed by coimmunoprecipitation as previously described [78]. 300 μg of gradient-purified mitochondria were centrifuged at 12,000 xg at 4°C for 10 min. The pellets were resuspended in 50 μl of lysis buffer (1% (w/v) digitonin, 50 mM NaCl, 2 mM EDTA, pH 8.0, 1mM PMSF, 10 mM HEPES, pH 7.4) and lysed by gentle agitation at 4°C for 15 min. 1 ml of washing buffer (0.1% (w/v) digitonin, 50 mM NaCl, 2 mM EDTA, pH 8.0, 1mM PMSF, 10 mM HEPES, pH 7.4) was then added, and samples were centrifuged at 15 min x 16,000 xg, 4°C. 40 μl of the supernatants were mixed with 8 μl of 6× Laemmli buffer, incubated for 10 min at 85°C and used as a loading control for the Western blot. The rest of the supernatants (960 μl) were mixed with 40 μl of Myc-Tag (9B11) magnetic beads (Cell Signaling) and incubated for 24 hours at 4°C with gentle rotation. After incubation, the beads were precipitated by quick centrifugation at 15 sec x 15,000 xg, and a 40 μl aliquot of the supernatant was mixed with 8 μl of 6× Laemmli buffer, incubated for 10 min at 85oC and used as an unbound control for Western blotting. The beads were washed three times with washing buffer. Elution of coimmunoprecipitated fraction from the pelleted beads was performed in 40 μl of lysis buffer mixed with 8 μl of 6× Laemmli buffer at 72°C for 10 min with vigorous agitation. Samples were quickly centrifuged at 30 sec x 15,000 xg, and the supernatants were transferred to the new tubes to be used for Western blotting.
References
- A. Sickmann, J. Reinders, Y. Wagner, C. Joppich, R. Zahedi, H.E. Meyer, B. Schönfisch, I. Perschil, A. Chacinska, B. Guiard, P. Rehling, N. Pfanner, and C. Meisinger, "The proteome of Saccharomyces cerevisiae mitochondria", Proceedings of the National Academy of Sciences, vol. 100, pp. 13207-13212, 2003. http://dx.doi.org/10.1073/pnas.2135385100
- K.N. Truscott, K. Brandner, and N. Pfanner, "Mechanisms of Protein Import into Mitochondria", Current Biology, vol. 13, pp. R326-R337, 2003. http://dx.doi.org/10.1016/s0960-9822(03)00239-2
- P. Dolezal, V. Likic, J. Tachezy, and T. Lithgow, "Evolution of the Molecular Machines for Protein Import into Mitochondria", Science, vol. 313, pp. 314-318, 2006. http://dx.doi.org/10.1126/science.1127895
- A.B. Taylor, B.S. Smith, S. Kitada, K. Kojima, H. Miyaura, Z. Otwinowski, A. Ito, and J. Deisenhofer, "Crystal Structures of Mitochondrial Processing Peptidase Reveal the Mode for Specific Cleavage of Import Signal Sequences", Structure, vol. 9, pp. 615-625, 2001. http://dx.doi.org/10.1016/S0969-2126(01)00621-9
- M. Yang, R.E. Jensen, M.P. Yaffe, W. Oppliger, and G. Schatz, "Import of proteins into yeast mitochondria: the purified matrix processing protease contains two subunits which are encoded by the nuclear MAS1 and MAS2 genes.", The EMBO journal, 1988. http://www.ncbi.nlm.nih.gov/pubmed/2905264
- M.P. Yaffe, and G. Schatz, "Two nuclear mutations that block mitochondrial protein import in yeast.", Proceedings of the National Academy of Sciences of the United States of America, 1984. http://www.ncbi.nlm.nih.gov/pubmed/6235522
- B.J. Smith, and M.P. Yaffe, "A mutation in the yeast heat-shock factor gene causes temperature-sensitive defects in both mitochondrial protein import and the cell cycle.", Molecular and cellular biology, 1991. http://www.ncbi.nlm.nih.gov/pubmed/2017170
- D.P. Atencio, and M.P. Yaffe, "MAS5, a yeast homolog of DnaJ involved in mitochondrial protein import.", Molecular and cellular biology, 1992. http://www.ncbi.nlm.nih.gov/pubmed/1729605
- M.K. Greene, K. Maskos, and S.J. Landry, "Role of the J-domain in the cooperation of Hsp40 with Hsp70.", Proceedings of the National Academy of Sciences of the United States of America, 1998. http://www.ncbi.nlm.nih.gov/pubmed/9600925
- A.J. Caplan, J. Tsai, P.J. Casey, and M.G. Douglas, "Farnesylation of YDJ1p is required for function at elevated growth temperatures in Saccharomyces cerevisiae.", The Journal of biological chemistry, 1992. http://www.ncbi.nlm.nih.gov/pubmed/1527016
- J. Li, X. Qian, and B. Sha, "The Crystal Structure of the Yeast Hsp40 Ydj1 Complexed with Its Peptide Substrate", Structure, vol. 11, pp. 1475-1483, 2003. http://dx.doi.org/10.1016/j.str.2003.10.012
- Z. Lu, and D.M. Cyr, "The Conserved Carboxyl Terminus and Zinc Finger-like Domain of the Co-chaperone Ydj1 Assist Hsp70 in Protein Folding", Journal of Biological Chemistry, vol. 273, pp. 5970-5978, 1998. http://dx.doi.org/10.1074/jbc.273.10.5970
- A.J. Caplan, and M.G. Douglas, "Characterization of YDJ1: a yeast homologue of the bacterial dnaJ protein.", The Journal of cell biology, vol. 114, pp. 609-621, 1991. http://dx.doi.org/10.1083/jcb.114.4.609
- A.J. Caplan, D.M. Cyr, and M.G. Douglas, "YDJ1p facilitates polypeptide translocation across different intracellular membranes by a conserved mechanism", Cell, vol. 71, pp. 1143-1155, 1992. http://dx.doi.org/10.1016/S0092-8674(05)80063-7
- F. Odds, "Candida and Candidosis", Bailliere Tindall, London, United Kingdom, 1988.
- D. Horn, D. Neofytos, E. Anaissie, J. Fishman, W. Steinbach, A. Olyaei, K. Marr, M. Pfaller, C. Chang, and K. Webster, "Epidemiology and Outcomes of Candidemia in 2019 Patients: Data from the Prospective Antifungal Therapy Alliance Registry", Clinical Infectious Diseases, vol. 48, pp. 1695-1703, 2009. http://dx.doi.org/10.1086/599039
- A.J.P. Brown, S. Budge, D. Kaloriti, A. Tillmann, M.D. Jacobsen, Z. Yin, I.V. Ene, I. Bohovych, D. Sandai, S. Kastora, J. Potrykus, E.R. Ballou, D.S. Childers, S. Shahana, and M.D. Leach, "Stress adaptation in a pathogenic fungus", Journal of Experimental Biology, vol. 217, pp. 144-155, 2014. http://dx.doi.org/10.1242/jeb.088930
- N.A.R. Gow, A.J.P. Brown, and F.C. Odds, "Fungal morphogenesis and host invasion.", Current opinion in microbiology, 2002. http://www.ncbi.nlm.nih.gov/pubmed/12160854
- M.D. Leach, and L.E. Cowen, "To Sense or Die: Mechanisms of Temperature Sensing in Fungal Pathogens", Current Fungal Infection Reports, vol. 8, pp. 185-191, 2014. http://dx.doi.org/10.1007/s12281-014-0182-1
- S. Nicholls, M.D. Leach, C.L. Priest, and A.J.P. Brown, "Role of the heat shock transcription factor, Hsf1, in a major fungal pathogen that is obligately associated with warm‐blooded animals", Molecular Microbiology, vol. 74, pp. 844-861, 2009. http://dx.doi.org/10.1111/j.1365-2958.2009.06883.x
- M.D. Leach, K.M. Tyc, A.J.P. Brown, and E. Klipp, "Modelling the Regulation of Thermal Adaptation in Candida albicans, a Major Fungal Pathogen of Humans", PLoS ONE, vol. 7, pp. e32467, 2012. http://dx.doi.org/10.1371/journal.pone.0032467
- M.D. Leach, E. Klipp, L.E. Cowen, and A.J.P. Brown, "Fungal Hsp90: a biological transistor that tunes cellular outputs to thermal inputs", Nature Reviews Microbiology, vol. 10, pp. 693-704, 2012. http://dx.doi.org/10.1038/nrmicro2875
- S. Diezmann, M.D. Leach, and L.E. Cowen, "Functional Divergence of Hsp90 Genetic Interactions in Biofilm and Planktonic Cellular States", PLOS ONE, vol. 10, pp. e0137947, 2015. http://dx.doi.org/10.1371/journal.pone.0137947
- S. Diezmann, M. Michaut, R.S. Shapiro, G.D. Bader, and L.E. Cowen, "Mapping the Hsp90 Genetic Interaction Network in Candida albicans Reveals Environmental Contingency and Rewired Circuitry", PLoS Genetics, vol. 8, pp. e1002562, 2012. http://dx.doi.org/10.1371/journal.pgen.1002562
- M.D. Leach, S. Budge, L. Walker, C. Munro, L.E. Cowen, and A.J.P. Brown, "Hsp90 Orchestrates Transcriptional Regulation by Hsf1 and Cell Wall Remodelling by MAPK Signalling during Thermal Adaptation in a Pathogenic Yeast", PLoS Pathogens, vol. 8, pp. e1003069, 2012. http://dx.doi.org/10.1371/journal.ppat.1003069
- M.D. Leach, R.A. Farrer, K. Tan, Z. Miao, L.A. Walker, C.A. Cuomo, R.T. Wheeler, A.J.P. Brown, K.H. Wong, and L.E. Cowen, "Hsf1 and Hsp90 orchestrate temperature-dependent global transcriptional remodelling and chromatin architecture in Candida albicans", Nature Communications, vol. 7, 2016. http://dx.doi.org/10.1038/ncomms11704
- M. Shingu-Vazquez, and A. Traven, "Mitochondria and Fungal Pathogenesis: Drug Tolerance, Virulence, and Potential for Antifungal Therapy", Eukaryotic Cell, vol. 10, pp. 1376-1383, 2011. http://dx.doi.org/10.1128/EC.05184-11
- M.J. Dagley, I.E. Gentle, T.H. Beilharz, F.A. Pettolino, J.T. Djordjevic, T.L. Lo, N. Uwamahoro, T. Rupasinghe, D.L. Tull, M. McConville, C. Beaurepaire, A. Nantel, T. Lithgow, A.P. Mitchell, and A. Traven, "Cell wall integrity is linked to mitochondria and phospholipid homeostasis in Candida albicans through the activity of the post‐transcriptional regulator Ccr4‐Pop2", Molecular Microbiology, vol. 79, pp. 968-989, 2010. http://dx.doi.org/10.1111/j.1365-2958.2010.07503.x
- I. Bohovych, S. Kastora, S. Christianson, D. Topil, H. Kim, T. Fangman, Y.J. Zhou, A. Barrientos, J. Lee, A.J.P. Brown, and O. Khalimonchuk, "Oma1 Links Mitochondrial Protein Quality Control and TOR Signaling To Modulate Physiological Plasticity and Cellular Stress Responses", Molecular and Cellular Biology, vol. 36, pp. 2300-2312, 2016. http://dx.doi.org/10.1128/MCB.00156-16
- A. Bambach, M.P. Fernandes, A. Ghosh, M. Kruppa, D. Alex, D. Li, W.A. Fonzi, N. Chauhan, N. Sun, O.A. Agrellos, A.E. Vercesi, R.J. Rolfes, and R. Calderone, "Goa1p of Candida albicans Localizes to the Mitochondria during Stress and Is Required for Mitochondrial Function and Virulence", Eukaryotic Cell, vol. 8, pp. 1706-1720, 2009. http://dx.doi.org/10.1128/EC.00066-09
- J.A. McDonough, V. Bhattacherjee, T. Sadlon, and M.K. Hostetter, "Involvement of Candida albicans NADH dehydrogenase complex I in filamentation", Fungal Genetics and Biology, vol. 36, pp. 117-127, 2002. http://dx.doi.org/10.1016/S1087-1845(02)00007-5
- N. Grahl, E.G. Demers, A.K. Lindsay, C.E. Harty, S.D. Willger, A.E. Piispanen, and D.A. Hogan, "Mitochondrial Activity and Cyr1 Are Key Regulators of Ras1 Activation of C. albicans Virulence Pathways", PLOS Pathogens, vol. 11, pp. e1005133, 2015. http://dx.doi.org/10.1371/journal.ppat.1005133
- J. Verghese, J. Abrams, Y. Wang, and K.A. Morano, "Biology of the Heat Shock Response and Protein Chaperones: Budding Yeast (Saccharomyces cerevisiae) as a Model System", Microbiology and Molecular Biology Reviews, vol. 76, pp. 115-158, 2012. http://dx.doi.org/10.1128/mmbr.05018-11
- S. Lindquist, and E.A. Craig, "THE HEAT-SHOCK PROTEINS", Annual Review of Genetics, vol. 22, pp. 631-677, 1988. http://dx.doi.org/10.1146/annurev.ge.22.120188.003215
- L.A. Walker, N.A. Gow, and C.A. Munro, "Fungal echinocandin resistance", Fungal Genetics and Biology, vol. 47, pp. 117-126, 2010. http://dx.doi.org/10.1016/j.fgb.2009.09.003
- N.A.R. Gow, F.L. van de Veerdonk, A.J.P. Brown, and M.G. Netea, "Candida albicans morphogenesis and host defence: discriminating invasion from colonization", Nature Reviews Microbiology, vol. 10, pp. 112-122, 2011. http://dx.doi.org/10.1038/nrmicro2711
- R.S. Shapiro, P. Uppuluri, A.K. Zaas, C. Collins, H. Senn, J.R. Perfect, J. Heitman, and L.E. Cowen, "Hsp90 Orchestrates Temperature-Dependent Candida albicans Morphogenesis via Ras1-PKA Signaling", Current Biology, vol. 19, pp. 621-629, 2009. http://dx.doi.org/10.1016/j.cub.2009.03.017
- F.L. Mayer, D. Wilson, I.D. Jacobsen, P. Miramón, S. Slesiona, I.M. Bohovych, A.J.P. Brown, and B. Hube, "Small but Crucial: The Novel Small Heat Shock Protein Hsp21 Mediates Stress Adaptation and Virulence in Candida albicans", PLoS ONE, vol. 7, pp. e38584, 2012. http://dx.doi.org/10.1371/journal.pone.0038584
- Y. Gong, Y. Kakihara, N. Krogan, J. Greenblatt, A. Emili, Z. Zhang, and W.A. Houry, "An atlas of chaperone–protein interactions in Saccharomyces cerevisiae : implications to protein folding pathways in the cell", Molecular Systems Biology, vol. 5, 2009. http://dx.doi.org/10.1038/msb.2009.26
- M. Babu, J. Vlasblom, S. Pu, X. Guo, C. Graham, B.D.M. Bean, H.E. Burston, F.J. Vizeacoumar, J. Snider, S. Phanse, V. Fong, Y.Y.C. Tam, M. Davey, O. Hnatshak, N. Bajaj, S. Chandran, T. Punna, C. Christopolous, V. Wong, A. Yu, G. Zhong, J. Li, I. Stagljar, E. Conibear, S.J. Wodak, A. Emili, and J.F. Greenblatt, "Interaction landscape of membrane-protein complexes in Saccharomyces cerevisiae", Nature, vol. 489, pp. 585-589, 2012. http://dx.doi.org/10.1038/nature11354
- J.R. Glover, and S. Lindquist, "Hsp104, Hsp70, and Hsp40: a novel chaperone system that rescues previously aggregated proteins.", Cell, 1998. http://www.ncbi.nlm.nih.gov/pubmed/9674429
- I. Gabriel, J. Olchowy, A. StanisÅawska-Sachadyn, T. Mio, J. Kur, and S. Milewski, "Phosphorylation of glucosamine-6-phosphate synthase is important but not essential for germination and mycelial growth ofCandida albicans", FEMS Microbiology Letters, vol. 235, pp. 73-80, 2004. http://dx.doi.org/10.1111/j.1574-6968.2004.tb09569.x
- T.R. O’Meara, A.O. Veri, T. Ketela, B. Jiang, T. Roemer, and L.E. Cowen, "Global analysis of fungal morphology exposes mechanisms of host cell escape", Nature Communications, vol. 6, 2015. http://dx.doi.org/10.1038/ncomms7741
- T. Roemer, B. Jiang, J. Davison, T. Ketela, K. Veillette, A. Breton, F. Tandia, A. Linteau, S. Sillaots, C. Marta, N. Martel, S. Veronneau, S. Lemieux, S. Kauffman, J. Becker, R. Storms, C. Boone, and H. Bussey, "Large‐scale essential gene identification in Candida albicans and applications to antifungal drug discovery", Molecular Microbiology, vol. 50, pp. 167-181, 2003. http://dx.doi.org/10.1046/j.1365-2958.2003.03697.x
- E. Taylor, and J. Rutter, "Mitochondrial quality control by the ubiquitin–proteasome system", Biochemical Society Transactions, vol. 39, pp. 1509-1513, 2011. http://dx.doi.org/10.1042/BST0391509
- V. Geli, M.J. Yang, K. Suda, A. Lustig, and G. Schatz, "The MAS-encoded processing protease of yeast mitochondria. Overproduction and characterization of its two nonidentical subunits.", The Journal of biological chemistry, 1990. http://www.ncbi.nlm.nih.gov/pubmed/2229072
- B.G. Oliver, P.M. Silver, C. Marie, S.J. Hoot, S.E. Leyde, and T.C. White, "Tetracycline alters drug susceptibility in Candida albicans and other pathogenic fungi", Microbiology, vol. 154, pp. 960-970, 2008. http://dx.doi.org/10.1099/mic.0.2007/013805-0
- N.D. Bonawitz, M.S. Rodeheffer, and G.S. Shadel, "Defective Mitochondrial Gene Expression Results in Reactive Oxygen Species-Mediated Inhibition of Respiration and Reduction of Yeast Life Span", Molecular and Cellular Biology, vol. 26, pp. 4818-4829, 2006. http://dx.doi.org/10.1128/MCB.02360-05
- B. Westermann, and W. Neupert, "Mitochondria-targeted green fluorescent proteins: convenient tools for the study of organelle biogenesis in Saccharomyces cerevisiae.", Yeast (Chichester, England), 2000. http://www.ncbi.nlm.nih.gov/pubmed/11054823
- K. MAHLKE, N. PFANNER, J. MARTIN, A.L. HORWICH, F. HARTL, and W. NEUPERT, "Sorting pathways of mitochondrial inner membrane proteins", European Journal of Biochemistry, vol. 192, pp. 551-555, 1990. http://dx.doi.org/10.1111/j.1432-1033.1990.tb19260.x
- M.E. Feder, and G.E. Hofmann, "HEAT-SHOCK PROTEINS, MOLECULAR CHAPERONES, AND THE STRESS RESPONSE: Evolutionary and Ecological Physiology", Annual Review of Physiology, vol. 61, pp. 243-282, 1999. http://dx.doi.org/10.1146/annurev.physiol.61.1.243
- M. Taipale, I. Krykbaeva, M. Koeva, C. Kayatekin, K. Westover, G. Karras, and S. Lindquist, "Quantitative Analysis of Hsp90-Client Interactions Reveals Principles of Substrate Recognition", Cell, vol. 150, pp. 987-1001, 2012. http://dx.doi.org/10.1016/j.cell.2012.06.047
- A.J. McClellan, Y. Xia, A.M. Deutschbauer, R.W. Davis, M. Gerstein, and J. Frydman, "Diverse Cellular Functions of the Hsp90 Molecular Chaperone Uncovered Using Systems Approaches", Cell, vol. 131, pp. 121-135, 2007. http://dx.doi.org/10.1016/j.cell.2007.07.036
- M. Taipale, G. Tucker, J. Peng, I. Krykbaeva, Z. Lin, B. Larsen, H. Choi, B. Berger, A. Gingras, and S. Lindquist, "A Quantitative Chaperone Interaction Network Reveals the Architecture of Cellular Protein Homeostasis Pathways", Cell, vol. 158, pp. 434-448, 2014. http://dx.doi.org/10.1016/j.cell.2014.05.039
- L.E. Cowen, and S. Lindquist, "Hsp90 Potentiates the Rapid Evolution of New Traits: Drug Resistance in Diverse Fungi", Science, vol. 309, pp. 2185-2189, 2005. http://dx.doi.org/10.1126/science.1118370
- A. Fiori, S. Kucharíková, G. Govaert, B.P.A. Cammue, K. Thevissen, and P. Van Dijck, "The Heat-Induced Molecular Disaggregase Hsp104 of Candida albicans Plays a Role in Biofilm Formation and Pathogenicity in a Worm Infection Model", Eukaryotic Cell, vol. 11, pp. 1012-1020, 2012. http://dx.doi.org/10.1128/EC.00147-12
- J.N. Sun, N.V. Solis, Q.T. Phan, J.S. Bajwa, H. Kashleva, A. Thompson, Y. Liu, A. Dongari-Bagtzoglou, M. Edgerton, and S.G. Filler, "Host Cell Invasion and Virulence Mediated by Candida albicans Ssa1", PLoS Pathogens, vol. 6, pp. e1001181, 2010. http://dx.doi.org/10.1371/journal.ppat.1001181
-
D. Saraswat, R. Kumar, T. Pande, M. Edgerton, and P.J. Cullen, "Signalling mucin Msb2 Regulates adaptation to thermal stress in
C andida albicans", Molecular Microbiology, vol. 100, pp. 425-441, 2016. http://dx.doi.org/10.1111/mmi.13326 - O. Gakh, P. Cavadini, and G. Isaya, "Mitochondrial processing peptidases", Biochimica et Biophysica Acta (BBA) - Molecular Cell Research, vol. 1592, pp. 63-77, 2002. http://dx.doi.org/10.1016/S0167-4889(02)00265-3
- R.J. Deshaies, B.D. Koch, M. Werner-Washburne, E.A. Craig, and R. Schekman, "A subfamily of stress proteins facilitates translocation of secretory and mitochondrial precursor polypeptides", Nature, vol. 332, pp. 800-805, 1988. http://dx.doi.org/10.1038/332800a0
- T. Endo, S. Mitsui, M. Nakai, and D. Roise, "Binding of mitochondrial presequences to yeast cytosolic heat shock protein 70 depends on the amphiphilicity of the presequence.", The Journal of biological chemistry, 1996. http://www.ncbi.nlm.nih.gov/pubmed/8626757
- N. Rowley, C. Prip-Buus, B. Westermann, C. Brown, E. Schwarz, B. Barrell, and W. Neupert, "Mdj1p, a novel chaperone of the DnaJ family, is involved in mitochondrial biogenesis and protein folding", Cell, vol. 77, pp. 249-259, 1994. http://dx.doi.org/10.1016/0092-8674(94)90317-4
- M. Schmitt, W. Neupert, and T. Langer, "Hsp78, a Clp homologue within mitochondria, can substitute for chaperone functions of mt-hsp70.", The EMBO journal, 1995. http://www.ncbi.nlm.nih.gov/pubmed/7628444
- A.J. Brown, M.D. Leach, and S. Nicholls, "The relevance of heat shock regulation in fungal pathogens of humans", Virulence, vol. 1, pp. 330-332, 2010. http://dx.doi.org/10.4161/viru.1.4.12364
- F. Sherman, "Getting started with yeast.", Methods in enzymology, 2002. http://www.ncbi.nlm.nih.gov/pubmed/12073320
- J. Shen, W. Guo, and J.R. Köhler, "CaNAT1 , a Heterologous Dominant Selectable Marker for Transformation of Candida albicans and Other Pathogenic Candida Species", Infection and Immunity, vol. 73, pp. 1239-1242, 2005. http://dx.doi.org/10.1128/IAI.73.2.1239-1242.2005
- M.D. Leach, and L.E. Cowen, "Membrane Fluidity and Temperature Sensing Are Coupled via Circuitry Comprised of Ole1, Rsp5, and Hsf1 in Candida albicans", Eukaryotic Cell, vol. 13, pp. 1077-1084, 2014. http://dx.doi.org/10.1128/ec.00138-14
- J. Lambert, J. Fillingham, M. Siahbazi, J. Greenblatt, K. Baetz, and D. Figeys, "Defining the budding yeast chromatin‐associated interactome", Molecular Systems Biology, vol. 6, 2010. http://dx.doi.org/10.1038/msb.2010.104
- G. Liu, J. Zhang, B. Larsen, C. Stark, A. Breitkreutz, Z. Lin, B. Breitkreutz, Y. Ding, K. Colwill, A. Pasculescu, T. Pawson, J.L. Wrana, A.I. Nesvizhskii, B. Raught, M. Tyers, and A. Gingras, "ProHits: integrated software for mass spectrometry–based interaction proteomics", Nature Biotechnology, vol. 28, pp. 1015-1017, 2010. http://dx.doi.org/10.1038/nbt1010-1015
- E.W. Deutsch, L. Mendoza, D. Shteynberg, T. Farrah, H. Lam, N. Tasman, Z. Sun, E. Nilsson, B. Pratt, B. Prazen, J.K. Eng, D.B. Martin, A.I. Nesvizhskii, and R. Aebersold, "A guided tour of the Trans‐Proteomic Pipeline", PROTEOMICS, vol. 10, pp. 1150-1159, 2010. http://dx.doi.org/10.1002/pmic.200900375
- D. Shteynberg, E.W. Deutsch, H. Lam, J.K. Eng, Z. Sun, N. Tasman, L. Mendoza, R.L. Moritz, R. Aebersold, and A.I. Nesvizhskii, "iProphet: Multi-level Integrative Analysis of Shotgun Proteomic Data Improves Peptide and Protein Identification Rates and Error Estimates", Molecular & Cellular Proteomics, vol. 10, pp. M111.007690, 2011. http://dx.doi.org/10.1074/mcp.M111.007690
- G. Teo, G. Liu, J. Zhang, A.I. Nesvizhskii, A. Gingras, and H. Choi, "SAINTexpress: Improvements and additional features in Significance Analysis of INTeractome software", Journal of Proteomics, vol. 100, pp. 37-43, 2014. http://dx.doi.org/10.1016/j.jprot.2013.10.023
- J.D.R. Knight, G. Liu, J.P. Zhang, A. Pasculescu, H. Choi, and A. Gingras, "A web‐tool for visualizing quantitative protein–protein interaction data", PROTEOMICS, vol. 15, pp. 1432-1436, 2015. http://dx.doi.org/10.1002/pmic.201400429
- O. Khalimonchuk, M. Jeong, T. Watts, E. Ferris, and D.R. Winge, "Selective Oma1 Protease-mediated Proteolysis of Cox1 Subunit of Cytochrome Oxidase in Assembly Mutants", Journal of Biological Chemistry, vol. 287, pp. 7289-7300, 2012. http://dx.doi.org/10.1074/jbc.M111.313148
- G. Daum, P.C. Böhni, and G. Schatz, "Import of proteins into mitochondria. Cytochrome b2 and cytochrome c peroxidase are located in the intermembrane space of yeast mitochondria.", The Journal of biological chemistry, 1982. http://www.ncbi.nlm.nih.gov/pubmed/6290489
- O. Khalimonchuk, M. Ott, S. Funes, K. Ostermann, G. Rödel, and J.M. Herrmann, "Sequential Processing of a Mitochondrial Tandem Protein: Insights into Protein Import in Schizosaccharomyces pombe", Eukaryotic Cell, vol. 5, pp. 997-1006, 2006. http://dx.doi.org/10.1128/EC.00092-06
- Y. Fujiki, A.L. Hubbard, S. Fowler, and P.B. Lazarow, "Isolation of intracellular membranes by means of sodium carbonate treatment: application to endoplasmic reticulum.", The Journal of cell biology, 1982. http://www.ncbi.nlm.nih.gov/pubmed/7068762
- J.M. Herrmann, and B. Westermann, "Analysis of Protein–Protein Interactions in Mitochondria", Mitochondria, 2nd Edition, pp. 743-759, 2007. http://dx.doi.org/10.1016/S0091-679X(06)80034-8
ACKNOWLEDGMENTS
We thank Zhen-Yuan Lin for help in the preparation of the AP-MS samples, and Cathy Collins for technical assistance. MDL is supported by a Sir Henry Wellcome Postdoctoral Fellowship (Wellcome Trust 096072), LEC is supported by a Canada Research Chair in Microbial Genomics and Infectious Disease and by Cana-dian Institutes of Health Research (CIHR) Grants MOP-119520 and MOP-86452. OK is supported by National Insti-tutes of Health grant 5R01GM108975. A-CG is supported by a CIHR Foundation Grant (FDN143301), Genome Cana-da Genomics Innovation Network (GIN) Node and Tech-nical Development Grants, and a Canada Research Chair in Functional Proteomics. J-PL was supported by a TD Bank Health Research Fellowship at the Lunenfeld-Tanenbaum Research Institute and by a Scholarship for the Next Gen-eration of Scientists from the Cancer Research Society. JLX is supported by a CIHR – Frederick Banting and Charles Best Canada Graduate Scholarship. The funding agencies had no role in the study design, data collection and inter-pretation, or the decision to submit the work for publication.
COPYRIGHT
© 2017

Ydj1 governs fungal morphogenesis and stress response, and facilitates mitochondrial protein import via Mas1 and Mas2 by Xie et al. is licensed under a Creative Commons Attribution 4.0 International License.