Back to article: Placeholder factors in ribosome biogenesis: please, pave my way
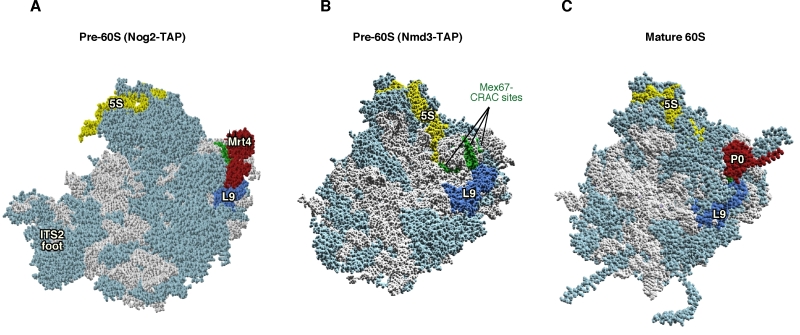
Figure 1: Mrt4 and Mex67 act as placeholder factors for the P0 r-protein. (A) Position of Mrt4 (red) in the early pre-60S r-particles purified with Nog2-TAP (PDB ID: 3JCT; [40]). (B) Mex67-binding sites at the P0 neighbourhood (green), identified by CRAC [42], have been highlighted in the late/cytoplasmic Nmd3-TAP pre-60S r-particle (PDB ID: 5H4P; [43]. (C) Position of P0 (red) in the mature 60S r-subunit (PDB ID: 3U5I, 3U5H; [44]). Particles are viewed from the subunit interface slightly turned to the left. For orientation, the positions of the 5S rRNA (yellow), the L9 r-protein (royal blue) and the above CRAC sites of Mex67 (green) have been highlighted in the three structures. The ITS2 foot has also labelled in A. Note that some of the CRAC sites of Mex67 overlap with Mrt4 and P0 in A and C, respectively. The rest of rRNAs are coloured in pale blue and the rest of r-proteins and/or factors in light cornflower blue. Images were generated using the UCSF Chimera program (www.cgl.ucsf.edu/chimera).
40. Wu S, Tutuncuoglu B, Yan K, Brown H, Zhang Y, Tan D, Gamalinda M, Yuan Y, Li Z, Jakovljevic J, Ma C, Lei J, Dong MQ, Woolford JL, Jr., Gao N (2016). Diverse roles of assembly factors revealed by structures of late nuclear pre-60S ribosomes. Nature 534(7605): 133-137. https://doi.org/10.1038/nature17942
42. Sarkar A, Pech M, Thoms M, Beckmann R, Hurt E (2016). Ribosome-stalk biogenesis is coupled with recruitment of nuclear-export factor to the nascent 60S subunit. Nat Struct Mol Biol 23(12): 1074-1082. https://doi.org/10.1038/nsmb.3312
43. Ma C, Wu S, Li N, Chen Y, Yan K, Li Z, Zheng L, Lei J, Woolford JL, Jr., Gao N (2017). Structural snapshot of cytoplasmic pre-60S ribosomal particles bound by Nmd3, Lsg1, Tif6 and Reh1. Nat Struct Mol Biol 24(3): 214-220. https://doi.org/10.1038/nsmb.3364
44. Ben-Shem A, Garreau de Loubresse N, Melnikov S, Jenner L, Yusupova G, Yusupov M (2011). The structure of the eukaryotic ribosome at 3.0 Å resolution. Science 334(6062): 1524-1529. https://doi.org/10.1126/science.1212642