Back to article: Aminoglycoside resistance profile and structural architecture of the aminoglycoside acetyltransferase AAC(6’)-Im
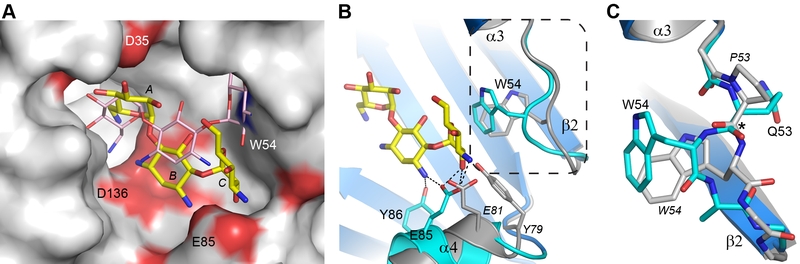
FIGURE 4: Structural comparison of AAC(6’)-Im and AAC(6’)-Ie. (A) Molecular surface representation of the binding site of AAC(6’)-Im (grey, red and blue shading). The kanamycin A molecule in AAC(6’)-Im binding site is shown as yellow sticks and the kanamycin A molecule in AAC(6’)-Ie is shown as thin pink sticks. (B) Ribbon representation of the loop between helix α3 and strand β2 shown in cyan for AAC(6’)-Im and grey for AAC(6’)-Ie. The location of the Trp54 side chain is indicated for both enzymes. The kanamycin A molecule in AAC(6’)-Im is shown as yellow sticks. The dashed box shows the area represented in panel C. (C) Close up view of the conformational difference caused by the presence of Pro53 in AAC(6’)-Ie (grey) relative to a Gln53 in AAC(6’)-Im (cyan). The difference in orientation of the residue 53 carbonyl group is indicated by the asterisk. Residues from AAC(6”)-Ie are labeled in italics.