Back to article: A versatile plasmid system for reconstitution and analysis of mammalian ubiquitination cascades in yeast
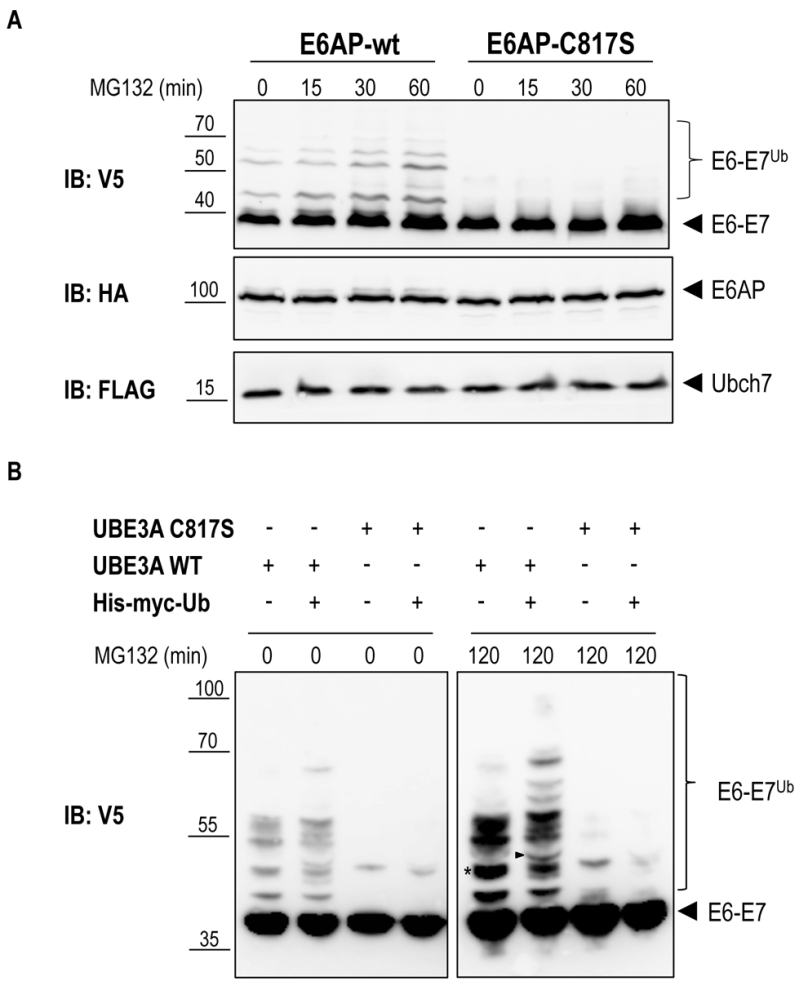
FIGURE 3: E6AP-dependent ubiquitination of E6-E7. (A) Yeast strain yRA2 was transformed with E6AP (wild type or catalytically inactive mutant) cloned in pRA306 (HA tag, LEU2 marker, TEF promoter), UBCH7 cloned in pRA300 (Flag tag, URA3 marker, TEF promoter) and E6-E7 cloned in pRA20 (V5 tag, TRP1 marker, 93%-GAL1 promoter). Following 1 hour of galactose induction, cells were treated with 75 μM MG132 for the indicated time points. Cells were lysed and equivalent amounts of protein extract were analyzed by SDS-PAGE and immunoblotting using the indicated antibodies. (B) Yeast strain yRA2 was transformed with E6AP (wild type or catalytically inactive mutant) cloned in pRA306 (HA tag, LEU2 marker, TEF promoter), E6-E7 cloned in pRA20 (V5 tag, TRP1 marker, 93%-GAL1 promoter) and 6xHis-Myc-Ub (Myc tag, HIS3 marker, CUP promoter) or empty p423 vector. Expression of 6xHis-Myc-Ub was induced by addition of CuSO4 (100 μM). Following 1 hour of galactose induction, cells were treated with 50 μM MG132 for 120 min. Cells were lysed and equivalent amounts of protein extract were analyzed by SDS-PAGE and immunoblotting using anti-V5 antibody. The band marked with an asterisk (*) is an example of an ubiquitinated E6-E7 band that in cells overexpressing 6xHis-Myc-Ub, generates additionally slower migrating species (arrow head) due to the incorporation of the 6xHis-Myc-tagged ubiquitin.