Back to article: Fat storage-inducing transmembrane (FIT or FITM) proteins are related to lipid phosphatase/phosphotransferase enzymes
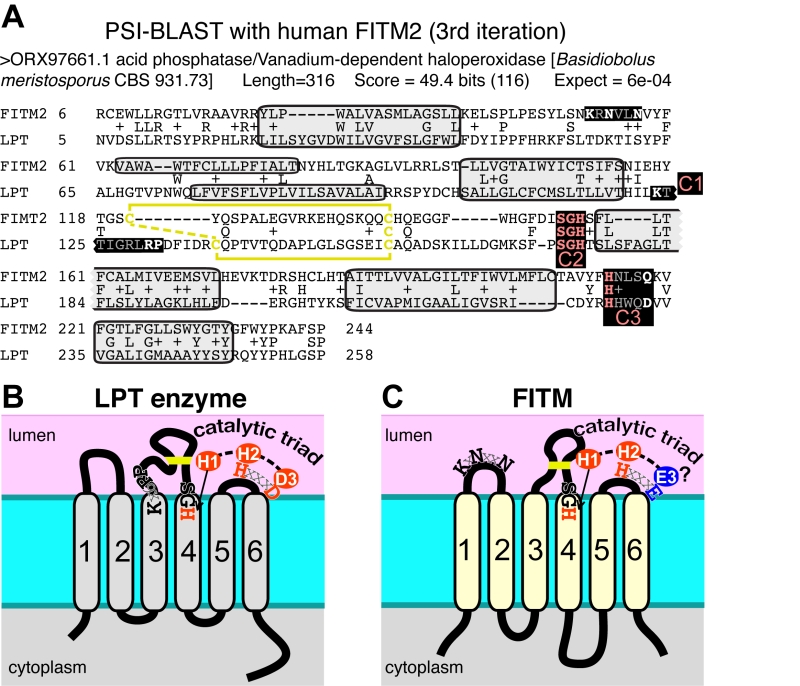
FIGURE 5: FITM proteins are related to lipid phosphatase/phosphotransferase (LPT) en-zymes. (A) The final significant hit (number 402) from PSI-BLAST with FITM2 after three iterations using the “nr50” database is a fungal LPT annotated as an acid phosphatase. The conserved residues of LPTs occur in three conserved motifs (C1 C2 C3) that are highlighted and colour coded: pink where precisely shared with FITM2 (C2: SGH, C3: HxxxD/E), white where not shared (C1: K-x6-RP in LPT only; KxNxxN in FITM2 only; C3: HxxxD/E). Variable positions within motifs are shown in gray. Predicted transmembrane domains (TMDs, shaded in light gray) align well in five cases. Paired cysteines likely to form a disulphide bridge in the loop between TMD3 and TMD4 are shown in yellow. (B) Topology of a typical LPT enzyme, Dpp1p in yeast, with the three conserved motifs, within which the catalytic triad of the most invariant residues are shown in red. (C) Topology of human FITM2, with the two motifs shared with LPTs and the KxNxxN motif. Residues that align with the catalytic triad are coloured red where identical with LPTs, and blue where non-identical.