Back to article: Fat storage-inducing transmembrane (FIT or FITM) proteins are related to lipid phosphatase/phosphotransferase enzymes
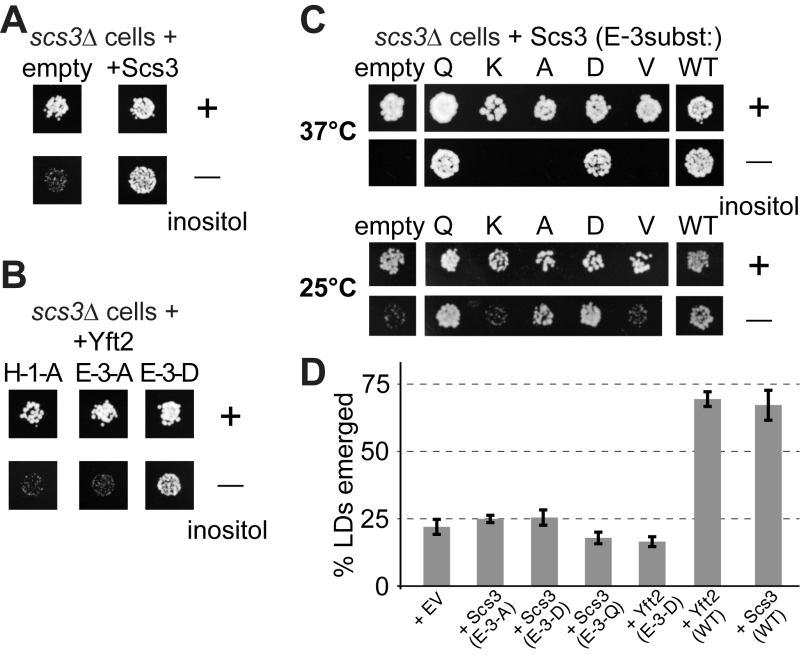
FIGURE 7: Growth of yeast lacking inositol requires FITMs with an intact catalytic triad. (A-C) Equal numbers of scs3∆ yeast (~50) transformed with the plasmids indicated were spotted onto plates either with added inositol (100 µM) or without, and grown at 30°C or the temperature indicated for 48-72 hours. (A) Cells either transformed with empty plasmid (expressing GFP alone) or expressing Scs3-GFP. (B) Cells grown in parallel to A, but expressing Yft2-GFP, either mutated H-1-A, E-3-A or E-3-D. (C) Cells were expressed with mutants of Scs3-GFP, substituting E-3 to five other residues. Here cells were grown at high stringency (37°C) or low stringency (25°C), the latter allowing rescue by the partially active construct E-3-A. (D) Emerged lipid droplets were counted in random sections of scs3∆ yft2∆ yeast expressing the indicated constructs (EV = empty vector), and identified as emerged from the ER by the complete absence of wrapping membrane (≥50 cells each experiment, n=3, SD shown by error bars). Emergence, as opposed to remaining wrapped in ER, was scored as previously [26]. This means that the droplets not counted here (i.e. not emerged) include both the clearly wrapped, and those with an ambiguous morphology (~7% under all conditions).
26. Choudhary V, Ojha N, Golden A, Prinz WA (2015). A conserved family of proteins facilitates nascent lipid droplet budding from the ER. J Cell Biol 211(2): 261-271. http://dx.doi.org/10.1083/jcb.201505067