Back to article: Snf1 cooperates with the CWI MAPK pathway to mediate the degradation of Med13 following oxidative stress
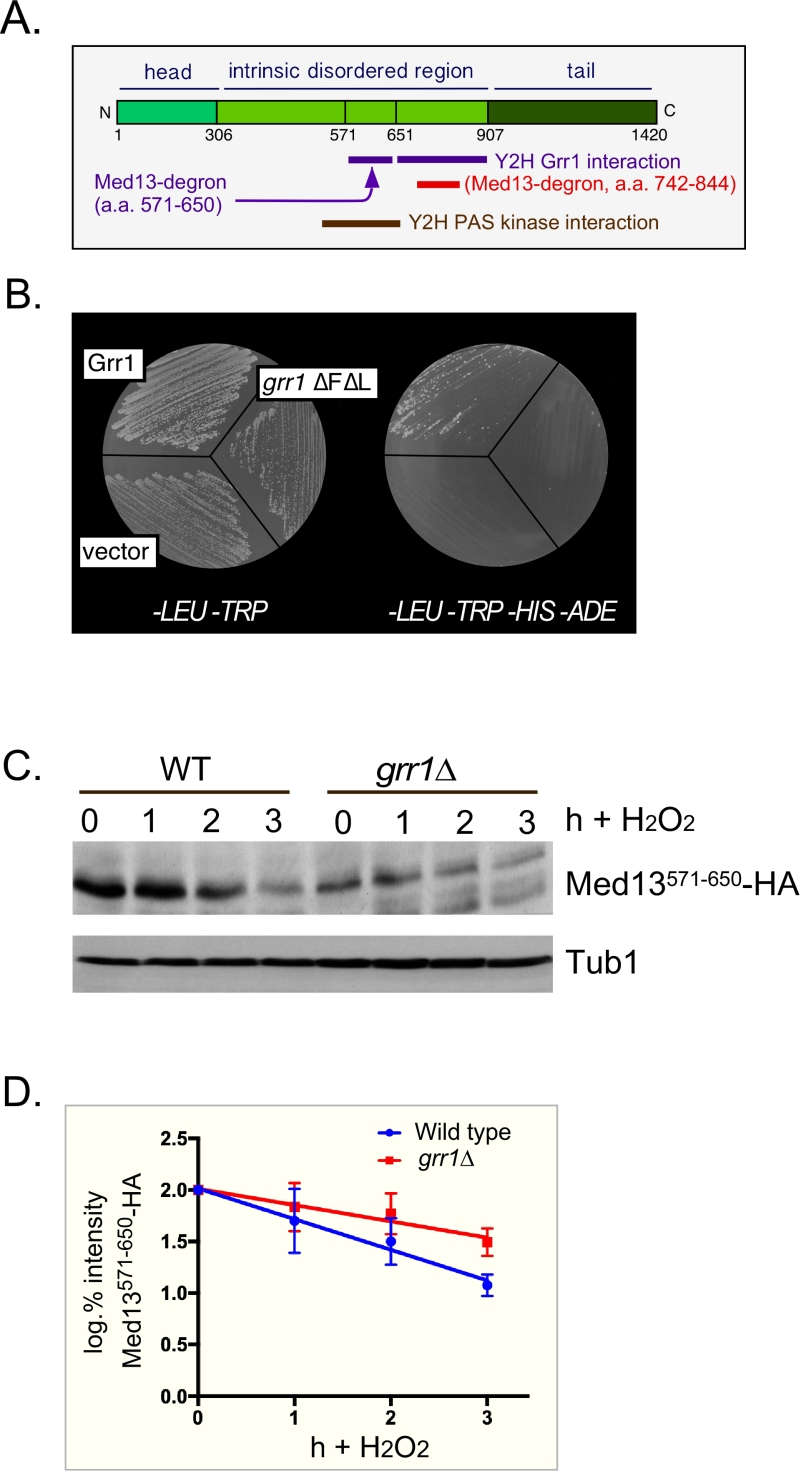
FIGURE 2: Med13 contains two H2O2 stress responsive degrons. (A) Cartoon of the results from ProteinPredict® [31] analysis of yeast Med13. The amino and carboxyl terminal domains are structured and separated by a large intrinsic disordered region. The positions of the two degrons are indicated. (B) Yeast two hybrid analysis of degron571-650 and Grr1 derivatives. Yeast PJ69-4a cells harboring Med13-activating domain plasmid (pDS15) and either pAS2, pAS-Grr1 or pAS2-Grr1∆F∆L binding domain plasmids were grown on -LEU, -TRP drop out medium to select for both plasmids (left panel) and -TRP, -LEU, -HIS –ADE (right panel) to test for Med13-Grr1 interaction. (C) Wild-type (RSY10) or grr1∆ cells (RSY1770) harboring degron571-650 (pDS15) were treated with 0.4 mM H2O2 for the timepoints indicated and Med13571-650-HA levels analyzed by Western blot. Tub1 levels were used as loading controls. (D) Degradation kinetics of the degron571-650 constructs shown in C. Values represent averages ± SD from a total of at least two Western blots from two independent experiments.
31. Yachdav G, Kloppmann E, Kajan L, Hecht M, Goldberg T, Hamp T, Honigschmid P, Schafferhans A, Roos M, Bernhofer M, Richter L, Ashkenazy H, Punta M, Schlessinger A, Bromberg Y, Schneider R, Vriend G, Sander C, Ben-Tal N, Rost B (2014). PredictProtein–an open resource for online prediction of protein structural and functional features. Nucleic Acids Res 42(Web Server issue): W337-343. http://dx.doi.org/10.1093/nar/gku366