Back to article: A chemical genetic screen reveals a role for proteostasis in capsule and biofilm formation by Cryptococcus neoformans
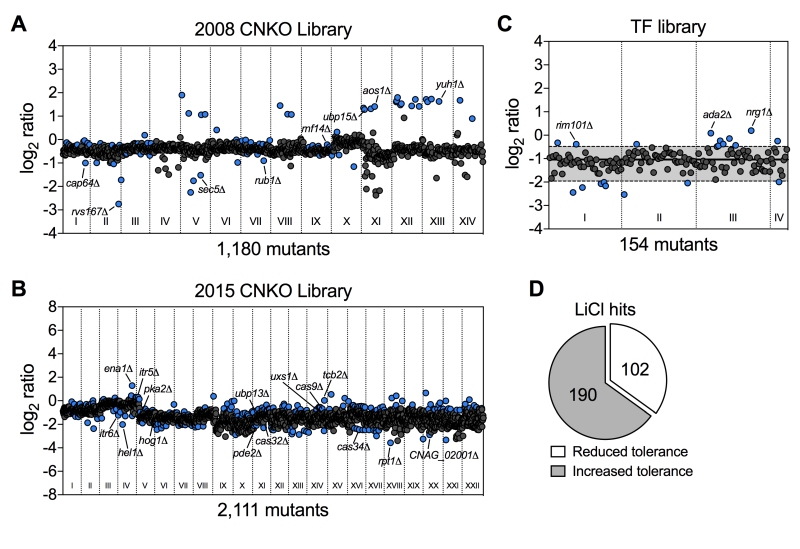
FIGURE 3: Chemical genetic analysis of the impact of lithium on C. neoformans. (A) Screening results for the 2008 C. neoformans knockout (CNKO) library. Results are based on OD600 measurements and are plotted as log2 ratio of growth in YPD medium supplemented with 100 mM lithium chloride relative to growth in YPD medium only. Roman numerals refer to the 96-well plate numbering as provided by the Fungal Genetics Stock Center (http://www.fgsc.net/). Mutants significantly affected by lithium were identified by calculating the mean log2-ratio of growth for all mutants of an individual plate and applying a cutoff of ± 1.5-fold the standard deviation (SD). Circles represent individual mutants. Blue colored circles indicate mutants that had significantly reduced (negative log2-ratios) or increased (positive log2-ratios) tolerance to lithium. Grey circles indicate mutants that did not show altered growth in presence of lithium. (B) Screening results for the 2015 CNKO library. See panel A for details. (C) Screening results for the transcription factor (TF) mutant library. See panel A for details. Note that for this smaller library, the mean and ± 1.5-fold SD cutoffs were calculated for mutants of all four plates combined instead of for individual plates. The mean is indicated by a solid black line, and the dashed lines indicate the 1.5-fold SD. (D) Proportion of mutants identified from the screen with reduced or increased tolerance towards lithium.