Back to article: Guidelines for DNA recombination and repair studies: Cellular assays of DNA repair pathways
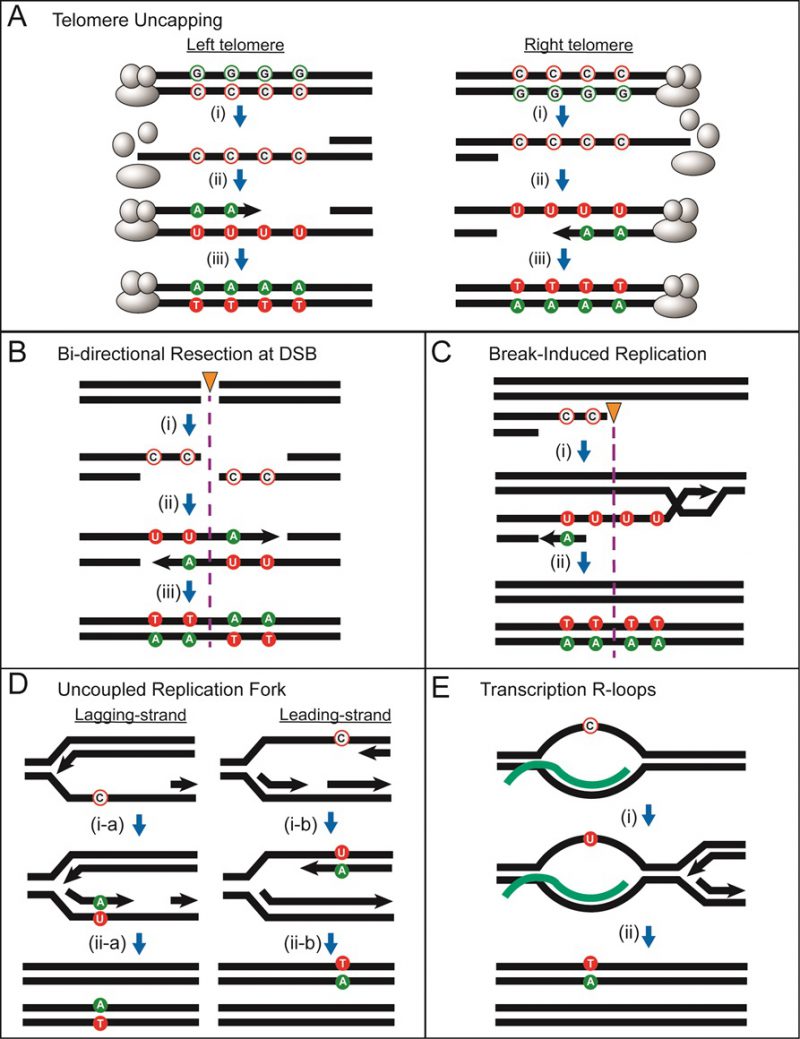
FIGURE 13: Cellular processes generating transient ssDNA vulnerable to hypermutation by APOBEC. The following symbols in the figure include: “unfilled green circles” = non-mutated guanines; “unfilled red circles” = non-mutated cytosines; “green-filled circles” = mutated guanines; “red-filled circles” = mutated cytosines; “black arrow heads” = 5′ to 3′ DNA synthesis; “orange triangle with purple dotted line” = position of double-strand break; “C” = cytosine; “A” = adenine; “U” = uracil; “T” = thymine. In all models, C to U changes are due to APOBEC cytosine deamination. (A) Long ssDNA generated at uncapped telomeres. (i) telomere uncapping triggers 5′ to 3′ end resection and G2 cell cycle arrest, generating long stretches of persistent ssDNA that provides a substrate for deamination of cytosines to uracils by APOBEC. (ii) the capping of telomeres is reestablished, initiating restoration of dsDNA with adenines inserted in the nascent DNA opposite the uracils. (iii) after the next round of replication, mutations are fixed, resulting in C:G to T:A transitions. Deamination of cytosines in the left telomeres result in G-coordinated clustered mutations on the top strand, while deamination of cytosines in the right telomeres result in C-coordinated clustered mutations on the top strand. (B) ssDNA formed from bi-directional resection at a double-strand break (DSB). (i) 5′ to 3′ end resection on both sides of a double strand break result in ssDNA in the top strand on the left-side of break and in ssDNA in the bottom strand on the right-side of the break. Cytosines in ssDNA on both sides of the break will be deaminated by APOBEC. (ii) and (iii) restoration of dsDNA using a sister chromatid or a homologous chromosome template followed by a round of replication will generate a single switch in strand coordination from C to G in a 5′→3′ direction of the top strand. (C) ssDNA formed from the repair of a one-ended DNA break via BIR. (i) 5′ to 3′ end resection results in a 3′ overhang that invades a homologous template and initiates DNA synthesis that progresses via a migrating replication bubble that generates a long ssDNA tail behind the bubble. Asynchronous lagging strand synthesis initiates to restore DNA to double strand form resulting in C-coordinated clustered mutations. If BIR initiated repair from the opposite side of a break such that ssDNA was generated on the bottom strand, then this would result in a G-coordinated cluster. (D) Long ssDNA could form from uncoupling between replicating leading and lagging strands. (i-a) and (ii-a) stalling of lagging strand synthesis resulting in ssDNA formed on the bottom strand leading to a mutated guanine. (i-b) and (ii-b) stalling of leading strand synthesis resulting in ssDNA formed in the top strand leading to a mutated cytosine. If uncoupling leads to long stretches of ssDNA, coordinated cluster formation is possible. In both cases, the origin of replication (not shown) is on the left side. The polarity of mutated cytosines or guanines depends on the fork direction. (E) Small stretches of ssDNA in the non-transcribed strand at transcription R-loops. (i) and (ii) ssDNA in the non-transcribed strand results in genome-wide bias towards APOBEC-mutated cytosines in non-transcribed strands. If uncoupling leads to long stretches of ssDNA, coordinated cluster formation is possible.