Back to article: Guidelines for DNA recombination and repair studies: Cellular assays of DNA repair pathways
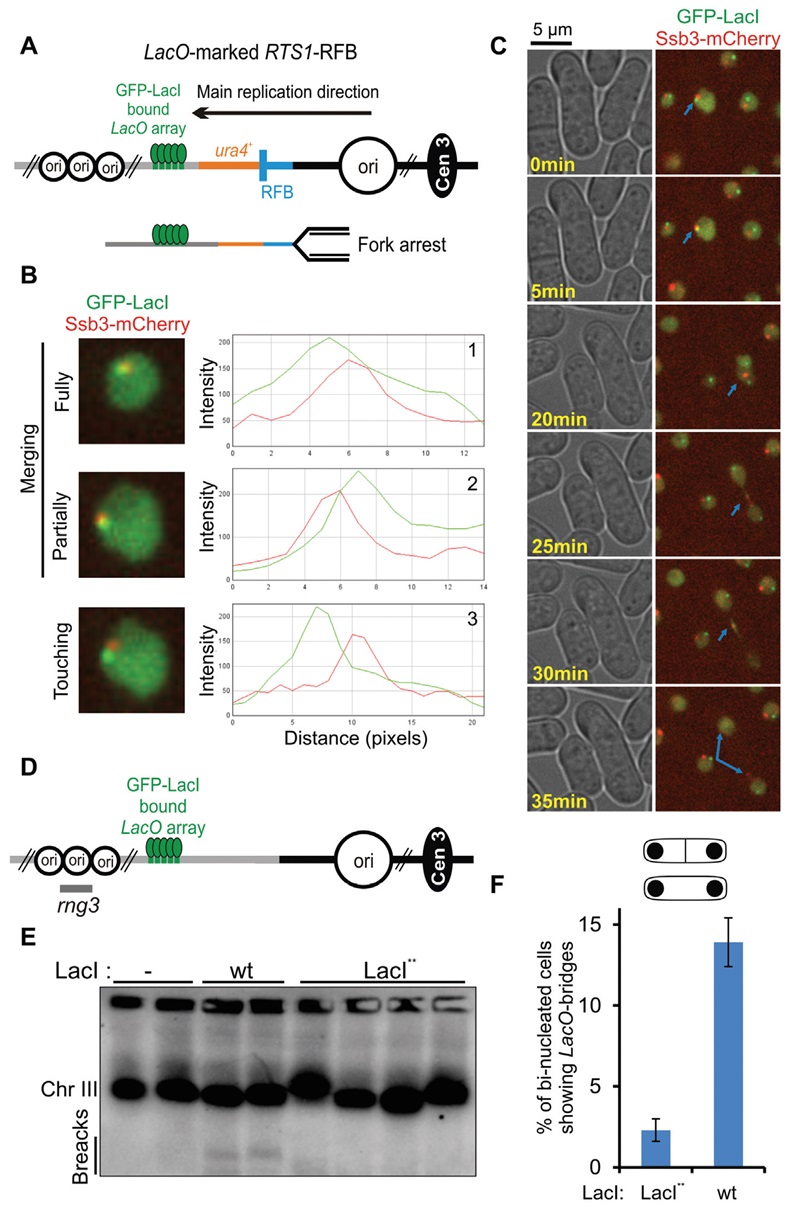
FIGURE 19: A fluorescently-marked RFB to follow in vivo the fate and processing of a single blocked fork in fission yeast. (A) Diagram of the LacO-marked RTS1-RFB locus located on the chromosome III of S. pombe. The blue bar indicates the RTS1-RFB and its polarity. Main replication origins (ori, black circles) located upstream and downstream from the RTS1-RFB are indicated. GFP-LacI (green ellipses) bound to LacO arrays (green bars) are integrated ∼7 kb away from the RTS1-RFB, on the telomere-proximal side of ura4 gene (red bar). When Rtf1 is expressed, >90% of forks emanating from the strong centromere-proximal replication origin, and moving towards the telomere, are blocked. (B) Example of Ssb3-mCherry foci being recruited to the active RFB. Three situations were considered: GFP-LacI and Ssb3-mCherry foci were fully (1) or partially merging (2), or they were touching each other (3). (C) Example of an unprotected fork (identified via the presence of Ssb3-mCherry-positive signal on LacO-marked RTS1-RFB in ON condition, blue arrow) in G2 (time point 0 to 20 minutes) converted into a sister chromatid bridge during mitotic progression (from 20 to 30 minutes) and finally resolved (at 35 minutes). (D) Diagram of the GFP-LacI bound to LacO arrays located on the chromosome III of S. pombe without RTS1-RFB. The rng3 probe is indicated. (E) Detection of chromosome breakage by Pulse Field Gel electrophoresis followed by Southern-blotting using the rng3 probe, in the following conditions: No LacI repressor expressed (-), expression of the wild-type (wt) LacI from a multi-copy expression vector 307], expression of the LacI** from a single copy gene and SV40 promoter [333]. (F) Quantification of LacO-bridges in indicated conditions.
307. Sofueva S, Osman F, Lorenz A, Steinacher R, Castagnetti S, Ledesma J, Whitby MC (2011). Ultrafine anaphase bridges, broken DNA and illegitimate recombination induced by a replication fork barrier. Nucleic Acids Res 39(15): 6568-6584. doi: 10.1093/nar/gkr340
333. Ait Saada A, Teixeira-Silva A, Iraqui I, Costes A, Hardy J, Paoletti G, Freon K, Lambert SAE (2017). Unprotected Replication Forks Are Converted into Mitotic Sister Chromatid Bridges. Mol Cell 66(3): 398-410 e394. doi: 10.1016/j.molcel.2017.04.002