Back to article: Guidelines for DNA recombination and repair studies: Mechanistic assays of DNA repair processes
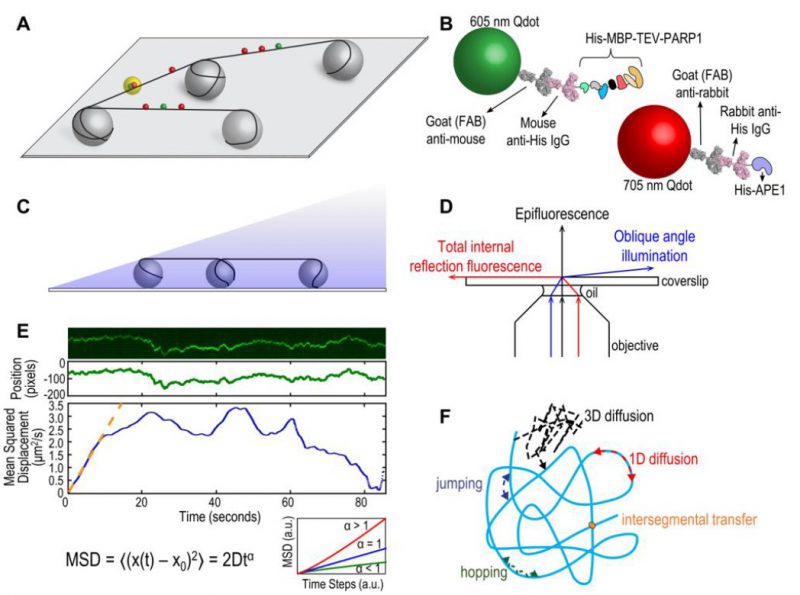
FIGURE 1: (A) Schematic of DNA tightrope setup. Long DNA molecules are suspended between 5 μm poly-L-lysine-coated silica beads on a glass coverslip. Qdot-labeled proteins bound to DNA shown in red and green (see B); colocalized particles highlighted in yellow. Adapted with permission from [13]. (B) Two orthogonal Qdot-protein labeling strategies. Top: A 605 nm Qdot (green) with conjugated anti-mouse secondary antibody (grey) bound to a mouse anti-His primary antibody (pink), bound to a His-tagged protein. Bottom: A 705 nm Qdot (red) with conjugated anti-rabbit secondary antibody (grey) bound to a rabbit anti-His primary antibody (pink), bound to a His-tagged protein. Adapted with permission from [13]. (C) DNA tightropes in a flow cell with oblique angle illumination. Adapted with permission from [8]. (D) Ray diagram showing incident laser light paths for epifluorescence (black), TIRF at the critical angle (red), and oblique angle illumination (blue). Adapted with permission from [8]. (E) Top: Sample kymograph of a Qdot-labeled protein displaying random linear diffusion on a DNA tightrope. Y axis, position; X axis, time. Middle: 1D Gaussian fittings of the light intensity profile at each time point of the above kymograph shown as position in pixels vs. time. 1 pixel = 46 nm. Bottom: Mean squared displacement (MSD) vs. time. The initial linear portion of the MSD plot is fit to the equation MSD = 2Dtα (orange line). Inset: Sub-types of 1D diffusion defined by α values. Super-diffusion (red), random diffusion (blue), sub-diffusion (green). Adapted with permission from[11]. (F) Modes of protein-DNA interaction. Search strategies typically involve some combination of: 3D diffusion in solution (black), 1D linear diffusion (red), jumping (blue) or hopping (green) between DNA segments, and intersegmental transfer (orange). Adapted with permission from [11].
8. Kong M, Beckwitt EC, Springall L, Kad NM, Van Houten B (2017). Single-Molecule Methods for Nucleotide Excision Repair: Building a System to Watch Repair in Real Time. Methods Enzymol 592: 213-257. 10.1016/bs.mie.2017.03.027
11. Kong M, Van Houten B (2017). Rad4 recognition-at-a-distance: Physical basis of conformation-specific anomalous diffusion of DNA repair proteins. Prog Biophys Mol Biol 127: 93-104. 10.1016/j.pbiomolbio.2016.12.004
13. Liu L, Kong M, Gassman NR, Freudenthal BD, Prasad R, Zhen S, Watkins SC, Wilson SH, Van Houten B (2017). PARP1 changes from three-dimensional DNA damage searching to one-dimensional diffusion after auto-PARylation or in the presence of APE1. Nucleic Acids Res 45(22): 12834-12847. 10.1093/nar/gkx1047