Back to article: Two TonB-dependent outer membrane transporters involved in heme uptake in Anabaena sp. PCC 7120
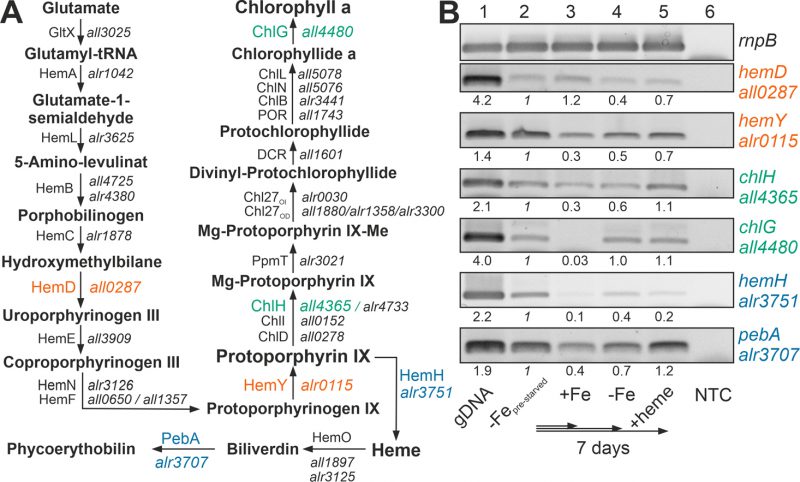
FIGURE 2: Expression of genes involved in chlorophyll a and heme synthesis after iron starvation or heme supply. (A) The synthesis pathway of heme and chlorophyll a, as well as the first steps of the heme degradation pathway, are shown. Intermediate metabolites, proteins involved and the gene coding for these proteins in Anabaena are indicated. Orange color indicates genes of the common pathway analyzed; green, genes analyzed of the chlorophyll a synthesis; blue, genes analyzed of the heme metabolism. HemF: O2-dependent coproporphyrinogen oxidase; HemN: O2-independent coproporphyrinogen dehydrogenase, CHL27OD/CHL27OI: aerobic/anaerobic magnesium-protoporphyrin IX monomethyl ester cyclase. Note: hemN and hemO are transcribed in a transcriptional unit; ChiL, ChiN and ChlB form a complex. (B) RNA was isolated from pre-starved Anabaena (lane 2) transferred for seven days to YBG11 (lane 3), YBG11 without iron (lane 4) or YBG11 without iron but 30 µM heme (lane 5). RT-PCR was performed using oligonucleotides for amplification of the indicated genes. The efficiency of the primer was controlled on isolated gDNA (lane 1), and RNA isolation was probed by RT-PCR without reverse transcriptase (NTC, lane 6). Amplification of rnpB is shown as control. The intensity of the bands shown was quantified with ImageJ, normalized to the expression of the genes in the pre-starved cultures and shown for guidance.