Back to article: Predictable regulation of survival by intratumoral microbe-immune crosstalk in patients with lung adenocarcinoma
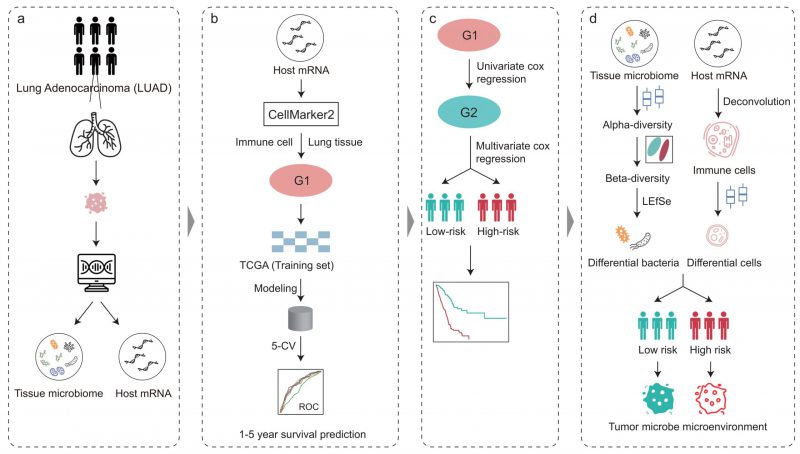
FIGURE 1: Overview of the analysis pipeline. The tumor microbiome abundance of LUAD was annotated by Poore et al. from RNA-Seq data and the matched host gene expression was downloaded from The Cancer Genome Atlas. Immune cell marker genes were used to build machine learning models to predict patient survival. COX regression analysis based on immune cell marker genes stratified patients into high- and low-risk. The intratumoral microbiota, the tumor immune microenvironment, and their crosstalk between the high-and low-risk groups were further explored.