Back to article: Replicative aging in yeast involves dynamic intron retention patterns associated with mRNA processing/export and protein ubiquitination
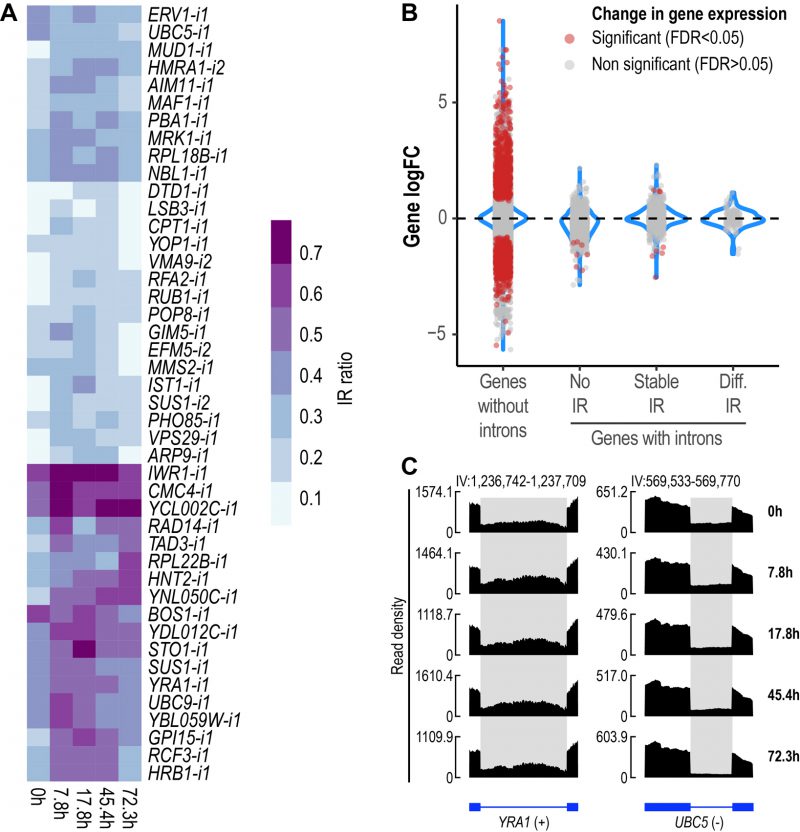
FIGURE 2: IR is altered during replicative yeast aging. (A) Heatmap displaying changes in IR levels of the 44 differentially retained introns identified during replicative yeast aging. (B) Distribution of the Log2 fold change values of genes without introns (single-exon genes) and genes with introns that show either no IR, stable IR levels or differential IR. Significant genes (False Discovery Rate (FDR) < 0.05) are shown in red. The plot includes the data of all the comparisons between the time points analyzed in this study. (C) RNA-seq tracks displaying changes in IR levels for YRA1 and UBC5. The y-axis shows read density. In the bottom part, the annotation of each gene is shown; exons are represented as boxes and introns as lines. Differentially retained introns are highlighted in grey. A plus (+) or a minus (-) sign next to the gene name indicates that the gene is encoded by the forward or reverse strand, respectively. RNA-seq tracks were generated with SparK [76].
76. Kurtenbach S, and Harbour JW (2019). SparK: A Publication-quality NGS Visualization Tool. bioRxiv 845529. 10.1101/845529