Back to article: Replicative aging in yeast involves dynamic intron retention patterns associated with mRNA processing/export and protein ubiquitination
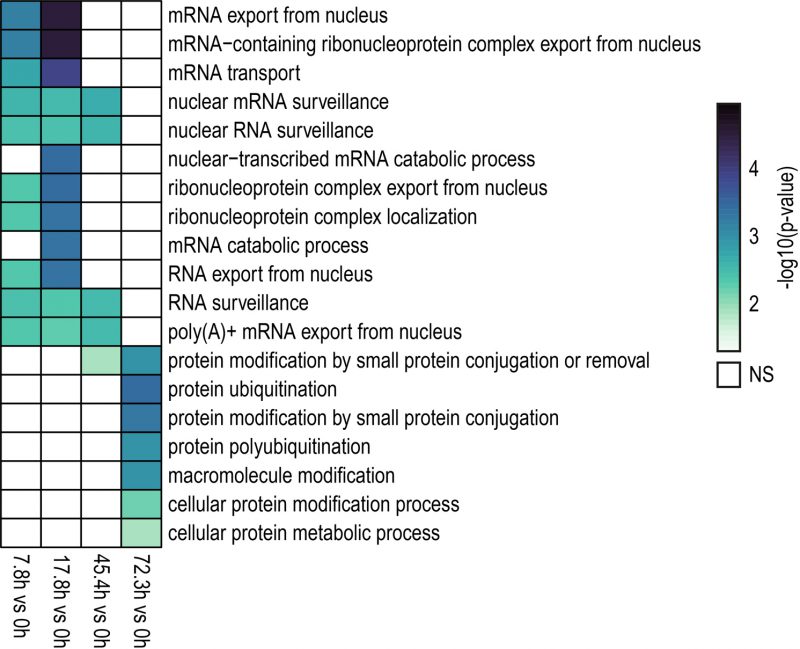
FIGURE 3: GO (Biological Process) analysis of genes showing differential IR levels during replicative yeast aging. Heatmap displaying the enriched biological processes identified for genes with differentially retained introns in the contrasts indicated in the bottom part (7.8 h vs 0 h, 17.8 h vs 0 h, 45.4 h vs 0 h, 72.3 h vs 0 h). Colored and blank boxes within the heatmap indicate significant and non-significant (NS) terms. The color scale represents the significance (Fisher's Exact test) of the term expressed as -log10 (p-value). GO terms with p < 0.05 were considered as significant. Note that during the early and middle stages of the replicative lifespan of yeast (7.8 h, 17.8 h and 45.4 h), genes with differentially retained introns were primarily involved in mRNA processing and export. On the other hand, genes exhibiting altered IR levels during the latest aging time point analyzed here (72.3 h) were mainly associated with protein ubiquitination.