Back to article: Polyadenylated versions of small non-coding RNAs in Saccharomyces cerevisiae are degraded by Rrp6p/Rrp47p independent of the core nuclear exosome
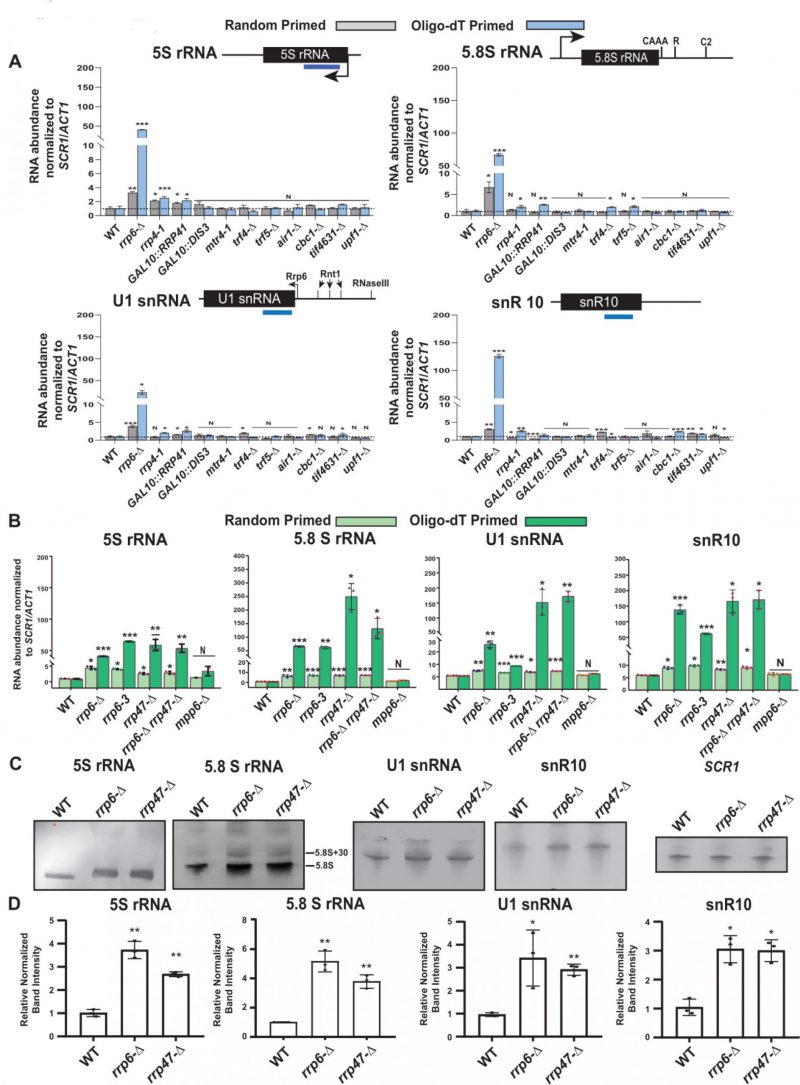
FIGURE 1: (A) Polyadenylated versions of small non-coding RNAs accumulate in an rrp6-Δ yeast strain. Histograms revealing the steady-state levels of 5S, 5.8S rRNAs, U1 and snR10 RNAs estimated from 2 ng cDNA samples prepared using either random hexanucleotide primers (grey bar) or oligo-dT30 anchor primer (blue bar) by RT-qPCR assay with the amplicons corresponding to their mature sequence from the indicated isogenic strains. The upf1-Δ strain was used as a negative control. (B-D) Characteristic steady-state levels of sncRNAs require the functional exonuclease domain of Rrp6p and ancillary nuclear factor Rrp47p/Lrp1p, but do not require Mpp6p. (B) Histogram revealing the steady-state levels of 5S, 5.8S, U1, and snR10 RNAs estimated by RT-qPCR from the 2 ng cDNA samples prepared using either random hexanucleotide primers (pale green bars) or oligo-dT30 anchor primer (deep green bars) from the indicated isogenic yeast strains. For histograms presented in panels A and B, SCR1 (in the case of Random Primer) and ACT1 mRNA (in the case of Oligo dT30 Primer) were used as the internal loading control. Normalized values of each of the ncRNAs in the WT yeast strain were set to one. Three to Four independent cDNA preparations (biological replicates, n = 3, in some cases 4) were used to determine the levels of various ncRNAs. The statistical significance of difference reflected in the ranges of P values estimated from Student’s two-tailed t-tests for a given pair of test strains for every message are presented with the following symbols, *<0.05, **<0.005, and ***<0.001; N, not significant. (C) Northern blots revealing the steady-state levels of 5S, 5.8S, U1, and snR10 RNAs in WT, rrp6-Δ and rrp47-Δ strains. Total RNA samples isolated from isogenic WT) and strains carrying rrp6-Δ, and rrp47-Δ yeast strains, separated on a 15% denaturing acrylamide gel and analyzed by northern blotting using DIG-labelled oligonucleotide probes corresponding to the mature regions of these rRNAs as described in materials and methods (See Supplementary Table S5). SCR1 RNA was used as a loading control. (D) Quantification of northern hybridization data for 5S, 5.8S, U1 and snR10 RNAs from panel B. Individual ncRNA levels were normalized to corresponding to SCR1 RNA signal. Normalized values of each rRNA in the WT yeast strain were set to one.