Back to article: Polyadenylated versions of small non-coding RNAs in Saccharomyces cerevisiae are degraded by Rrp6p/Rrp47p independent of the core nuclear exosome
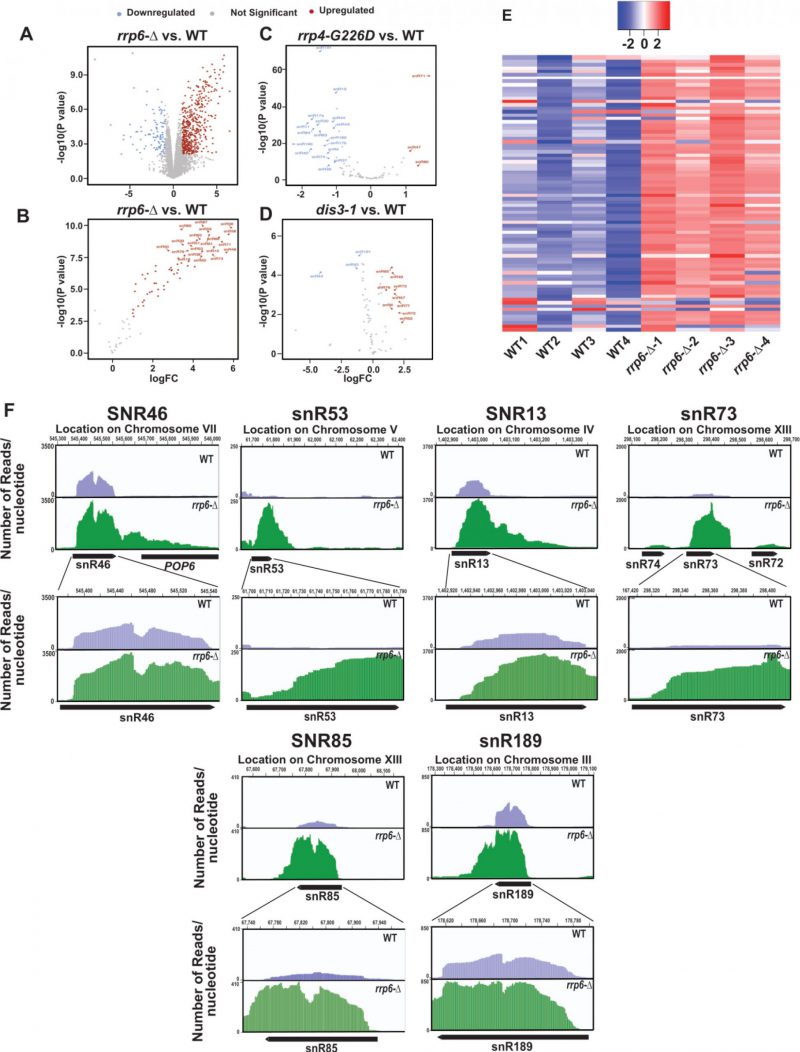
FIGURE 2: Analysis of previously published RNA-seq datasets (Accession Numbers, GSE135056, GSE163106, and GSE134295) revealed a dramatic accumulation of reads corresponding to the mature region of several small nucleolar RNAs. Volcano plots depicting the differential expression of all annotated sequences in rrp6-Δ (A), and all 82 sn- and snoRNA transcripts in rrp6-Δ (B), mutant rrp4-G226D (C), and mutant dis3-1 (D) yeast strain relative to WT strain. (E) Heat Map of normalized counts, showing the expression pattern for all 82 sn- and snoRNAs across four independent biological replicates of WT and rrp6-Δ yeast strains. (F) Graphical representation showing the relative amount of reads mapped to the genomic locus corresponding to small nucleolar RNAs snR46, snR53, snR13, snR73, snR71, snR685, and snR7189. The top panels depict the distant view (accommodating the 3′ extended regions), and the bottom panels show the close-up views for each snoRNA loci. The location of transcripts and the direction of transcription are shown below the graph (drawn in scale) by the solid black arrow-headed rectangles.