Back to article: Polyadenylated versions of small non-coding RNAs in Saccharomyces cerevisiae are degraded by Rrp6p/Rrp47p independent of the core nuclear exosome
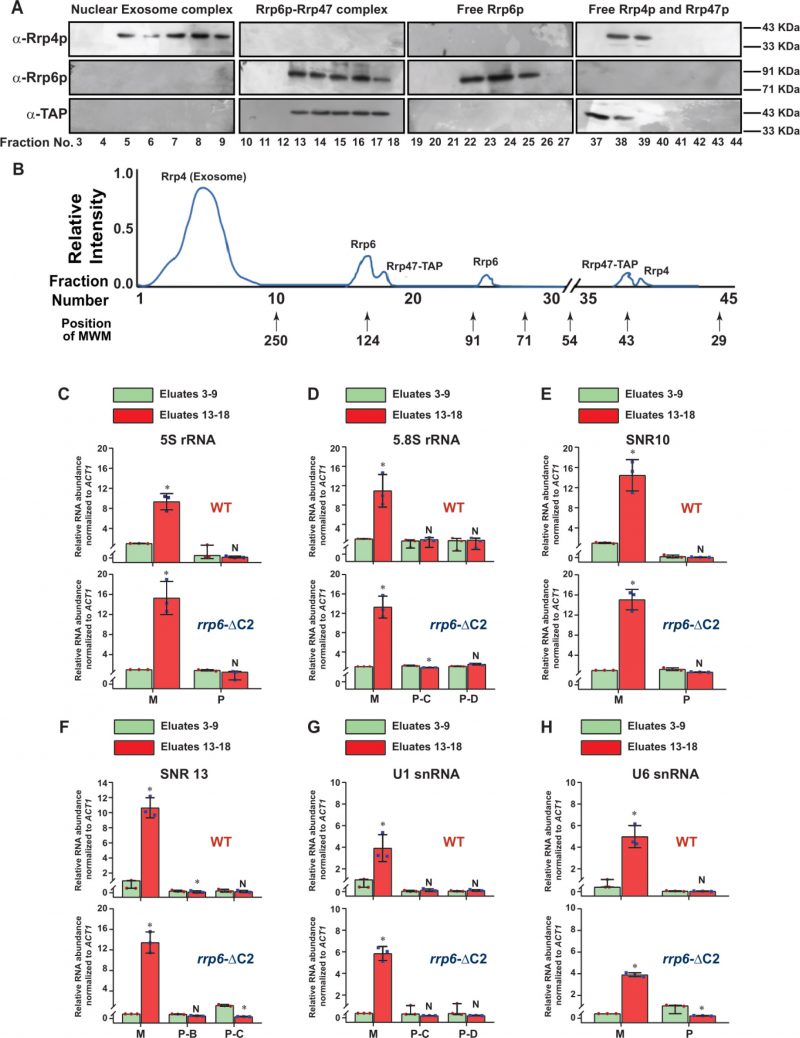
FIGURE 5: Rrp6p-ΔC2 and Rrp47p co-purify as a separate independent protein complex that cross-links to the entire population of ma-ture/processed species of sncRNAs. (A) Fractionation/separation profiles of the components of the nuclear exosome, RrpG-ΔC2/47p and free Rrp6p, Rrp4p, and Rrp47p in Biogel p-200 gel titration chromatographic column showing Rrp6p-AΔ2-Rrp47p exists as a separate complex independent of the core nuclear exosome. Growing cultures of yeast strains expressing Rrp6-AΔ2 and Rrp47-TAP (yBD-527) were subjected to UV irradiation for inducing RNA-protein cross-linking before harvesting as described in the materials and methods. Cell extract was subsequently prepared from the harvested cells that were further subjected to the fractionation by Biogel P-200 Gel filtration column as described in materials and methods and the eluted fractions were subjected to western blotting analysis using anti-Rrp4p, anti-Rrp6p, and anti-TAP (for detection of Rrp47p). Panel (B) depicts a qualitative representation of the elution profile of different proteins in the Biogel P-200 Gel filtration column. Note that in this experiment no Rrp6p/47p signal was detected in the fraction numbers 3–9 when they were either probed with anti-Rrp6p or anti-TAP antibodies showing a complete lack of interaction/association with the nuclear exosome. Relative elution volumes of various MW markers eluted from this column and the fractions in which they appeared are indicated in this profile. (C-H) Polyadenylated forms of all mature sncRNAs are enriched in the fractions that contain the Rrp6-AƊ2p-Rrp47p complex. Scattered/Bar plots revealing the distribution of the total (mature + precursor forms together) population and different precursor species of various low molecular weight ncRNAs in the pooled fractions 5–9 (corresponding to the position of the core nuclear exosome, green histograms) and 13–18 (corresponding to the position of the Rrp6/47p complex, orange histograms). Extracts were prepared from growing cultures of WT (yBD-507) and rrp6-ΔC2 (yBD-527) strains that were subjected to UV-crosslinking before harvesting. These cross-linked cell extracts were subjected to fractionation in the Biogel P-200 gel filtration column as before. Fraction numbers, 5–9 and 13–18 were subsequently pooled and subjected to RNA isolation and cDNA synthesis using oligo-dTjoanchor primer followed by RT-qPCR assay with 2 ng of cDNA using amplicons corresponding to CDS and 3′-extended precursor regions of various sncRNAs. ACT1 mRNA was used as the internal loading control. Normalized values of the total population of each ncRNA in both WT and rrp6-ΔC2 strains obtained from the pooled fractions 5–9 were set to 1. Three independent cDNA preparations (biological replicates, n = 3) were used to determine the levels of various ncRNAs. The statistical significance of difference reflected in the ranges of P values estimated from Student's two-tailed t-tests for a given pair of test strains for every message are presented with the following symbols, *<0.05, **<0.005, and ***<0.001; N, not significant.