Back to article: Polyadenylated versions of small non-coding RNAs in Saccharomyces cerevisiae are degraded by Rrp6p/Rrp47p independent of the core nuclear exosome
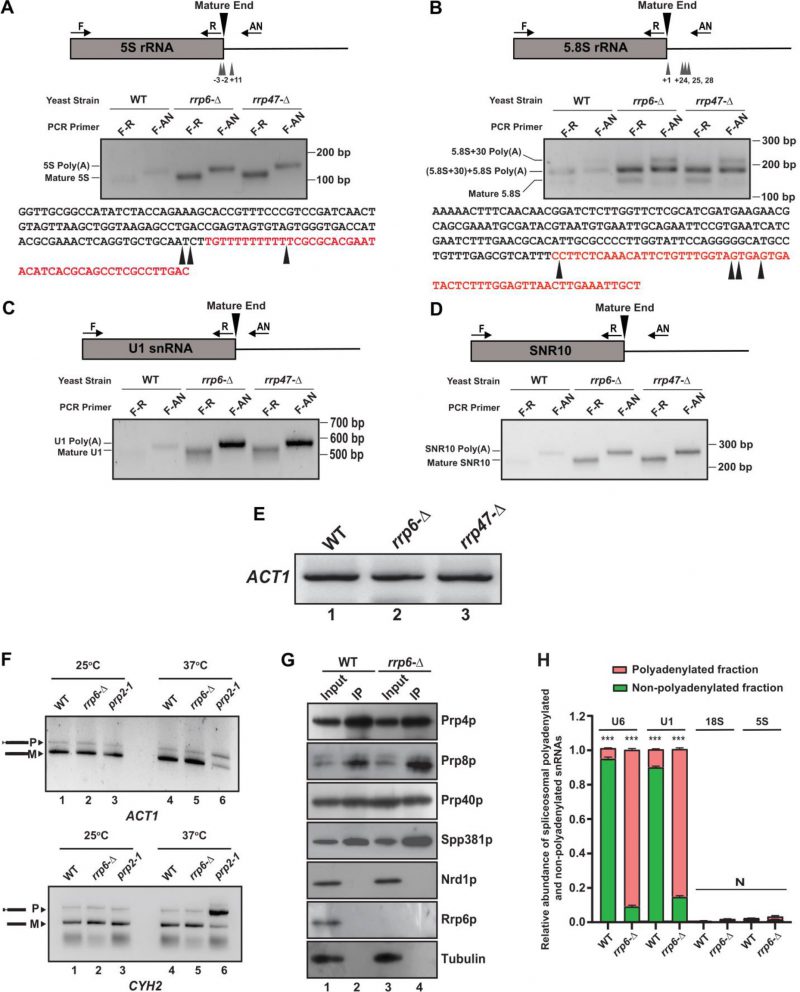
FIGURE 7: Polyadenylated versions of sncRNAs are accumulated in an rrp6-Δ mutant yeast strain. (A-B) Polyadenylation profiles of 5S (A), and 5.8S (B), RNAs. Each panel depicts a cartoon of the mature RNA along with the primer positions (F, R, and AN), the major polyadenylation sites. Gel images below the cartoon show the LM-PAT products of specific RNAs obtained with various primer sets at the bottom. The bottom parts of each panel show the genomic sequences of various ncRNAs with arrowheads indicating major polyadenylation sites identified. (C-D) Polyadenylation profiles of U1 (C), and snR10 (D) RNAs showing the cartoon of the mature RNA with the primer positions (F, R, and AN) and gel images depicting their LM-PAT products obtained with various primer sets. LM-PAT cDNA samples of these RNAs were prepared from isogenic WT (yBD-263) and strains carrying rrp6-Δ (yBD-265), and rrp47-Δ (yBD-454) yeast strains as described in materials and methods and then PCR amplified with either F-R (mature) and F-AN (polyadenylated) primer-sets specific for various RNAs followed by electrophoresis on a 2.5 – 3% agarose gel before photographed. (E) Relative levels of ACT1 mRNA amplified using ACT1 specific primer sets (see Table S3 for their sequence) from the LM-PAT cDNA prepared from WT (yBD-263), rrp6-Δ (yBD-265), and rrp47-Δ (yBD-454) yeast strains as described in materials and methods. (F) Deletion in RRP6, rrp6-Δ does not impair the splicing of the ACT1 and CYH2 pre-mRNAs at 37°C. Relative steady-state levels of precursor and mature forms of ACT1 and CYH2 mRNAs at 25°C and 37°C temperature in the isogenic WT (yBD-122), rrp6-Δ (yBD-70), and and prp2-1 (yBD-77) strains. Representative gel showing the relative levels of the precursor as well as mature ACT1 and CYH2 mRNA in these strains as determined by the semi-quantitative end-point PCR reactions carried out using the primer sets located within the exon1 and exon2 of each gene. Indicated strains were grown at 25°C till the OD of the culture reaches 0.6 followed by division of each culture into two halves. One half was continued to grow at 25°C and the other half was shifted to 37°C. Both cultures were continued to grow for 2 hour after the shift before harvesting. (G) Result from a representative co-immunoprecipitation experiment using hexa-HA-tagged Prp4p as bait to show purity of the spliceosomal prepration. Protein extracts from WT (yBD608) and rrp6-Δ (yBD614) cells expressing genomically hexa-HA-tagged version of Prp4p, were subjected to the immunopurification procedure. The immunoprecipitate (IP) was recovered, fractionated by electrophoresis on denaturing SDS gel followed by the detection of the specific spliceosomal and non-spliceosomal components using western blotting with specific antibodies as mentioned in Materials and Methods. The input (Lanes 1 and 3) in case of each sample is 100 µg of whole-cell protein extract from the respective strains, while the Co-IP (Lanes 2 and 4) for each lane represents 20 µl of the eluate in SDS-gel loading buffer from respective strains. (H) Relative abundance of polyadenylated and non-polyadenylated snRNAs in the immunopurified spliceosomal preparations. The immunoprecipitate (IP) from WT and rrp6-Δ yeast cells both expressing genomically tagged hexa-HA-tagged version of Prp4p was subjected to isolation of total RNA, followed by preparation of random-primed and oligo-dT primed cDNA and subsequent quantification of polyadenylated and non-polyadenylated fractions of U6/U1 snRNAs and 18S/5S rRNAs using 2 ng cDNA in each case. ACT1 mRNA were used as the internal loading control. Normalized values of combined fractions of adenylated and non-adenylated snRNAs in each strain were set to 1. Three independent cDNA preparations (biological replicates, n = 3) were used. P values of the adenylated fractions were estimated in each case with respect to the non-adenylated fractions, which were considered as reference with the following symbols, *<0.05, **<0.005, and ***<0.001; N, not significant.