Back to article: Polyadenylated versions of small non-coding RNAs in Saccharomyces cerevisiae are degraded by Rrp6p/Rrp47p independent of the core nuclear exosome
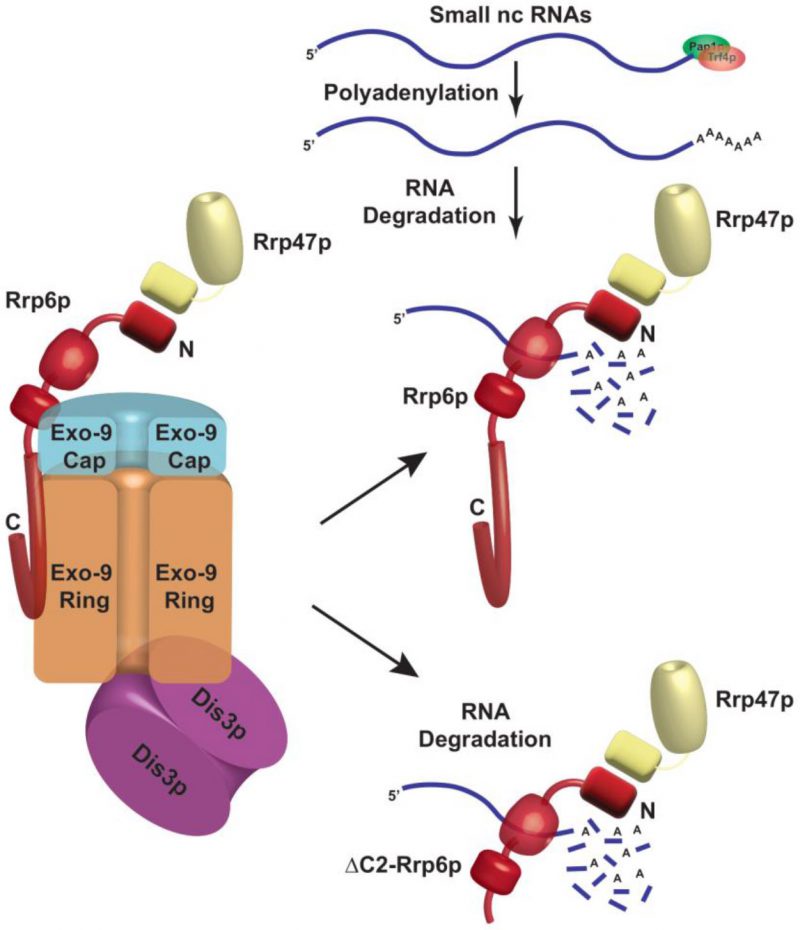
FIGURE 8: Cartoon showing a proposed model of the Rrp6p and Rrp47p-dependent degradation of polyadenylated mature/processed forms of sncRNAs in S. cerevisiae. Low molecular weight sncRNAs undergo polyadenylation by both the canonical and non-canonical poly(A) polymerases, Pap1p and Trf4p. This polyadenylated fraction of sncRNAs are then targeted by the Rrp6p-Rrp47p complex in the core-exosome independent manner. Notably, a C-terminally truncated version of Rrp6p that lacks the physical association with the core nuclear exosome is equally competent in targeting and degrading these sncRNAs.