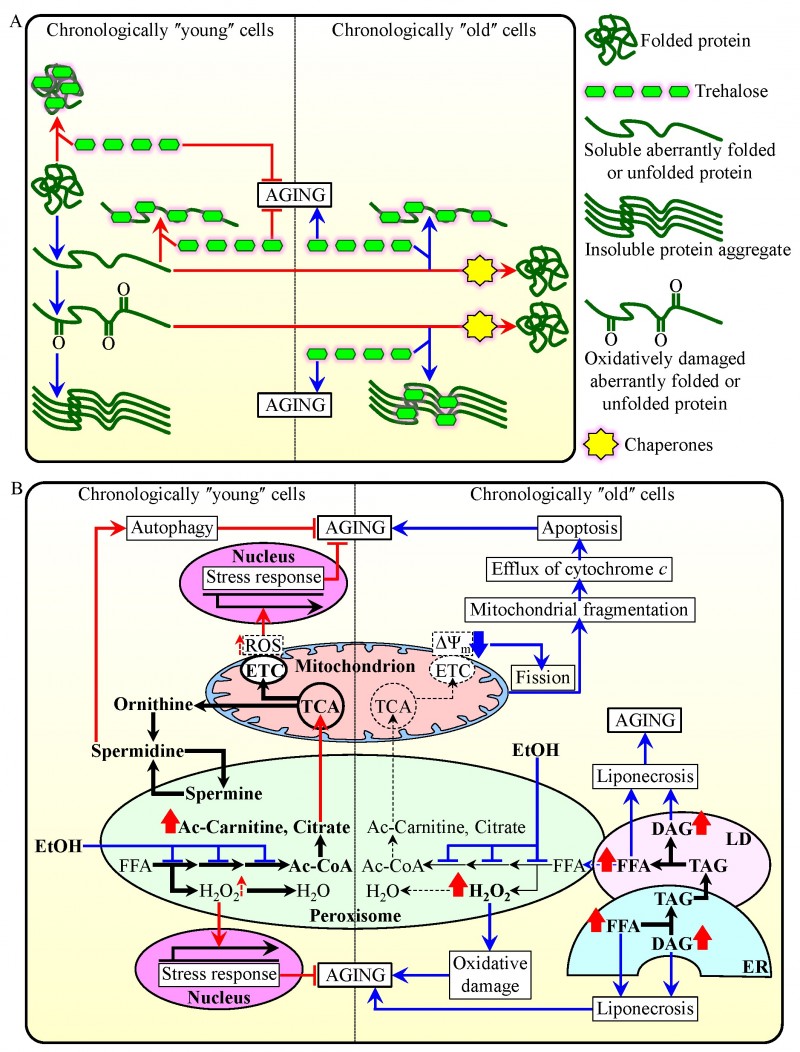
FIGURE 1: Cell-autonomous mechanisms define longevity of chronologically aging yeast by orchestrating trehalose metabolism and metabolic processes confined to peroxisomes.
(A) A model for molecular mechanisms through which trehalose regulates the process of cellular aging in yeast by modulating protein folding, misfolding, unfolding, refolding, oxidative damage, solubility and aggregation in chronologically ʺyoungʺ and ʺoldʺ cells.
(B) A model for how the age-dependent efficiency of peroxisomal protein import in chronologically aging yeast defines the age-related metabolic pattern of peroxisomes, thus impacting longevity-defining processes in other cellular compartments and ultimately establishing a pro- or anti-aging cellular pattern. Activation arrows and inhibition bars denote pro-aging processes (displayed in blue color) or anti-aging processes (displayed in red color). Please see text for additional details.
Ac-Carnitine, acetyl-carnitine; Ac-CoA, acetyl-CoA; DAG, diacylglycerol; ER, endoplasmic reticulum; ETC, electron transport chain; EtOH, ethanol; FFA, non-esterified (ʺfreeʺ) fatty acids; LD, lipid droplet; ROS, reactive oxygen species; TAG, triacylglycerols; TCA, tricarboxylic acid cycle; ΔΨm, electrochemical potential across the inner mitochondrial membrane.