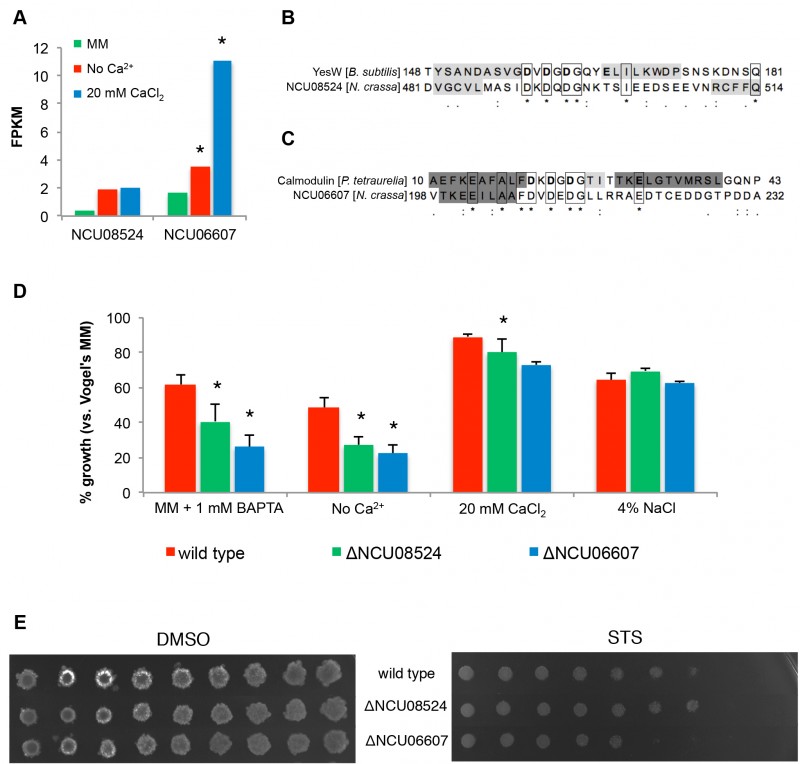
FIGURE 6: Identification of putative novel Ca2+-binding proteins.
(A) Expression levels of NCU08524 and NCU06607 in no Ca2+, standard MM and 20 mM CaCl2 media. *, p-value ≤ 0.05.
(B-C) Sequence alignments of the EF-hand-like domain of NCU08524 and YesW from B. subtilis (accession number: O31526; PDB code: 2z8r) (B), and NCU06607 and calmodulin from P. tetraurelia (accession number: P07463; PDB code: 1exr) (C). Residues binding Ca2+ in YesW and calmodulin are in bold; β-strands and α-helices are shaded in light and dark grey, respectively (confirmed by crystallography for YesW [36] and calmodulin [37] and predicted with MEMSAT3 for NCU08524 and NCU06607).
(D) The percentages of growth of the different strains in the indicated media versusstandard MM were determined by measuring absorbance of the respective liquid cultures at 450 nm. *, p-value ≤ 0.05. (E) The spot assay was used to examine the sensitivity of ΔNCU08524 and ΔNCU06607 to 5 μM staurosporine (STS).
36. Ochiai A, Itoh T, Maruyama Y, Kawamata A, Mikami B, Hashimoto W, Murata K (2007). A novel structural fold in polysaccharide lyases: Bacillus subtilis family 11 rhamnogalacturonan lyase YesW with an eight-bladed beta-propeller. J Biol Chem 282(51): 37134-37145. http://dx.doi.org/10.1074/jbc.M704663200
37. Wilson MA, Brunger AT (2000). The 1.0 A crystal structure of Ca2+-bound calmodulin: an analysis of disorder and implications for functionally relevant plasticity. J Mol Biol 301(5): 1237-1256. http://dx.doi.org/10.1006/jmbi.2000.4029