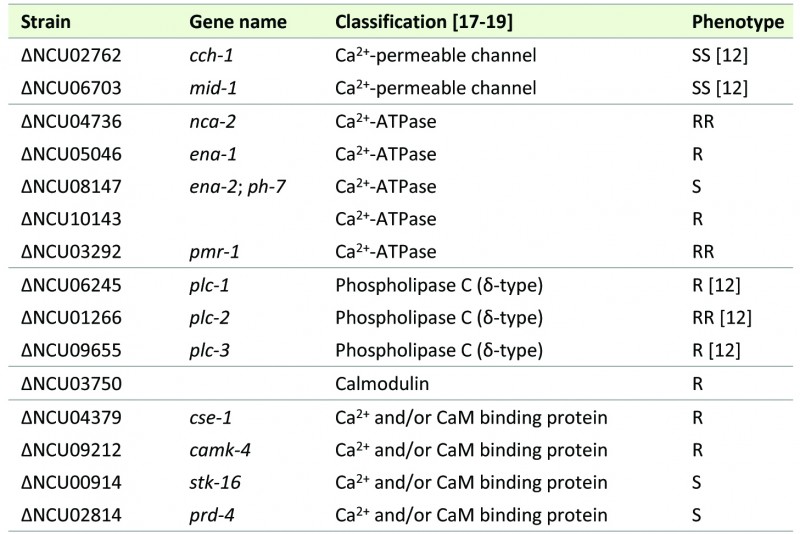
TABLE 1. Strains with Ca2+-related deleted genes that exhibit altered sensitivity to staurosporine when compared with wild type N. crassa.
SS: much more sensitive; S: slightly more sensitive; RR: much more resistant; R: slightly more resistant.
12. Gonçalves AP, Cordeiro JM, Monteiro J, Muñoz A, Correia-de-Sá P, Read ND, Videira A (2014). Activation of a TRP-like channel and intracellular calcium dynamics during phospholipase C-mediated cell death. J Cell Sci. (In press) http://dx.doi.org/10.1242/jcs.152058
17. Galagan JE, Calvo SE, Borkovich KA, Selker EU, Read ND, Jaffe D, FitzHugh W, Ma LJ, Smirnov S, Purcell S, Rehman B, Elkins T, Engels R, Wang S, Nielsen CB, Butler J, Endrizzi M, Qui D, Ianakiev P, Bell-Pedersen D, Nelson MA, Werner-Washburne M, Selitrennikoff CP, Kinsey JA, Braun EL, Zelter A, Schulte U, Kothe GO, Jedd G, Mewes W, et al. (2003). The genome sequence of the filamentous fungus Neurospora crassa. Nature 422(6934): 859-868. http://dx.doi.org/10.1038/nature01554
18. Borkovich KA, Alex LA, Yarden O, Freitag M, Turner GE, Read ND, Seiler S, Bell-Pedersen D, Paietta J, Plesofsky N, Plamann M, Goodrich-Tanrikulu M, Schulte U, Mannhaupt G, Nargang FE, Radford A, Selitrennikoff C, Galagan JE, Dunlap JC, Loros JJ, Catcheside D, Inoue H, Aramayo R, Polymenis M, Selker EU, Sachs MS, Marzluf GA, Paulsen I, Davis R, Ebbole DJ, et al.(2004). Lessons from the genome sequence of Neurospora crassa: tracing the path from genomic blueprint to multicellular organism. Microbiol Mol Biol Rev 68(1): 1-108. http://dx.doi.org/10.1128/MMBR.68.1.1-108.2004
19. Zelter A, Bencina M, Bowman BJ, Yarden O, Read ND (2004). A comparative genomic analysis of the calcium signaling machinery in Neurospora crassa, Magnaporthe grisea, and Saccharomyces cerevisiae. Fungal Genet Biol 41(9): 827-841. http://dx.doi.org/10.1016/j.fgb.2004.05.001