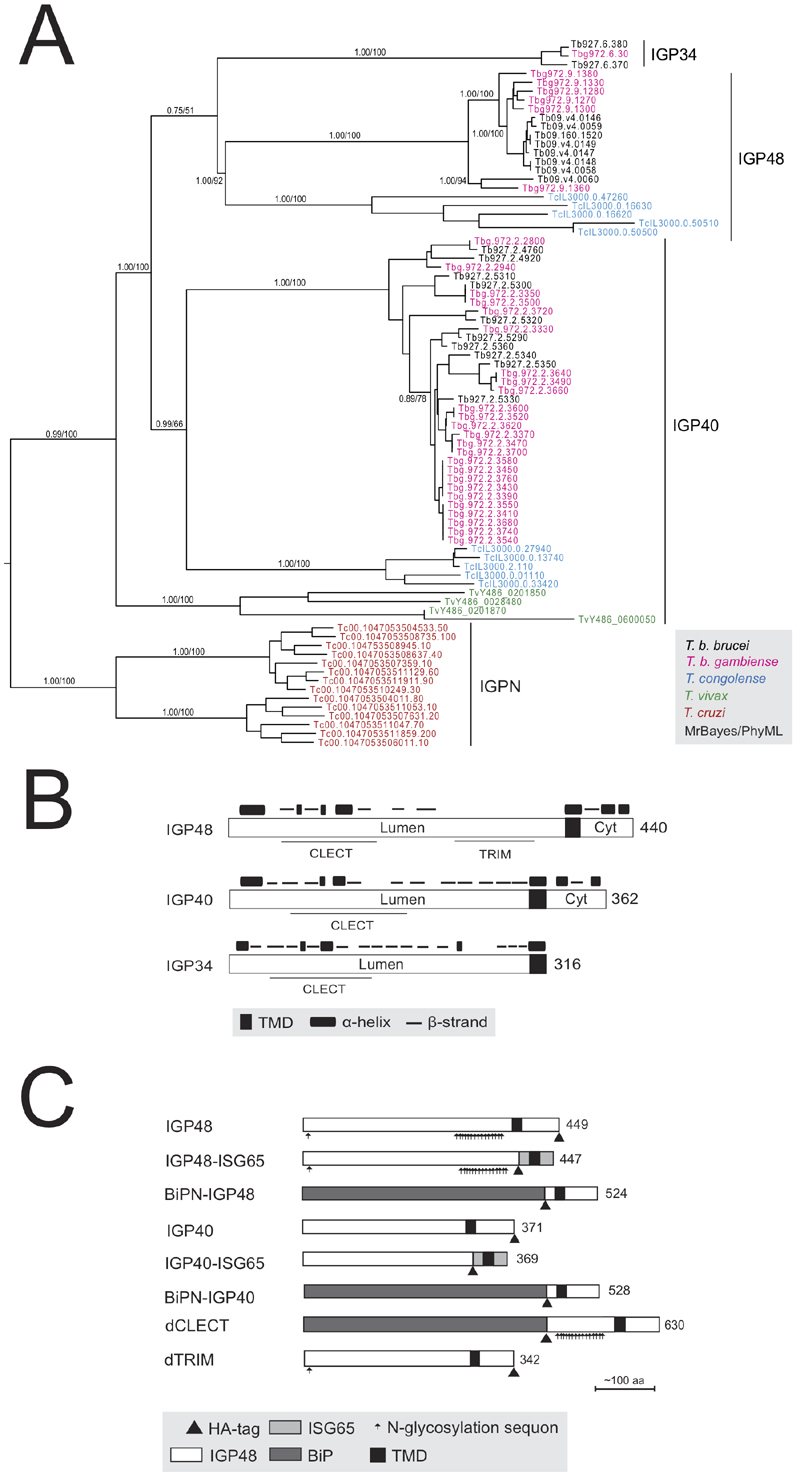
FIGURE 1: The invariant glycoprotein (IGP) family.
(A) Phylogenetic reconstruction of the IGP family. The tree shown is the best Bayesian topology with branch support for important nodes indicated from both Bayesian and PhyML calculations. Clades are indicated by vertical bars.
(B) Schematic diagram showing the secondary structure prediction and domain architecture of the IGP family. Lumen indicates the portion of the molecule predicted to be within the ER lumen and Cyt designates the predicted cytoplasmic domain. CLECT indicates C-type lectin domain.
(C) Schematic structures of epitope-tagged and domain-swap IGP constructs used in this study. All constructs contain an HA tag as indicated by small triangle below the bar.
Abbreviations:
IGP48, full length IGP48;
IGP48-ISG65, IGP48 ectodomain fused to ISG65 TMD and cytoplasmic domain;
BiPN-IGP48, IGP48 ectodomain replaced with BiPN, retaining IGP48 TMD and cytoplasmic domain;
IGP40, full length IGP40;
IGP40-ISG65, IGP40 ectodomain fused to ISG65 TMD and cytoplasmic domain;
BiPN-IGP40, IGP40 ectodomain replaced with BiPN, retaining IGP40 TMD and cytoplasmic domain;
dTRIM, IGP48 with predicted trimerisation domain deleted.
For figures in higher resolution please refer to PDF version of the article.