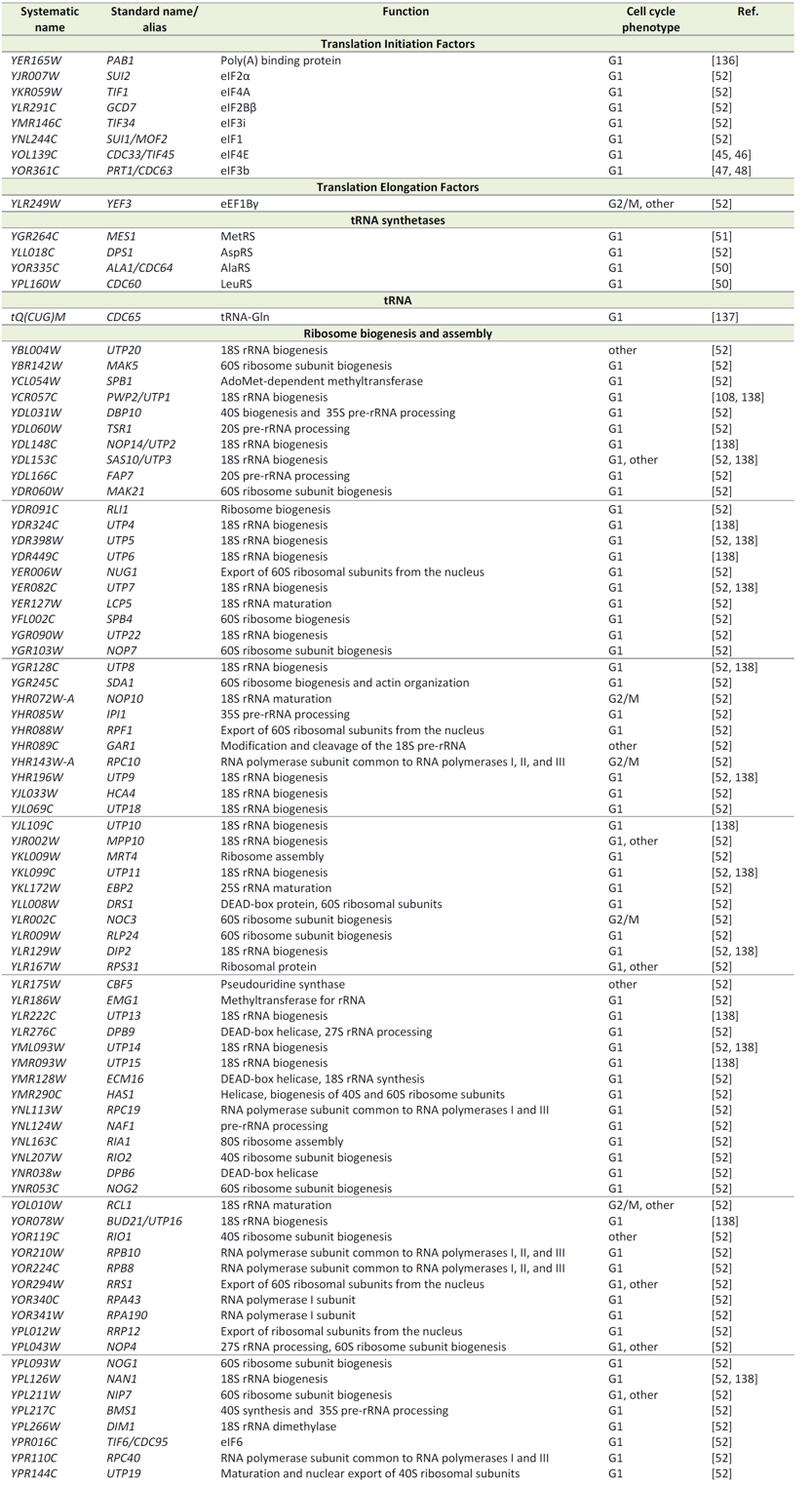
TABLE 1. Cell cycle phenotypes of loss-of-function mutants in essential genes encoding protein synthesis and ribosome biogenesis factors in S. cerevisiae.
45. Altmann M, Trachsel H (1989). Altered mRNA cap recognition activity of initiation factor 4E in the yeast cell cycle division mutant cdc33. Nucleic acids research 17(15): 5923-5931. http://dx.doi.org/10.1093/nar/17.15.5923
46. Brenner C, Nakayama N, Goebl M, Tanaka K, Toh-e A, Matsumoto K (1988). CDC33 encodes mRNA cap-binding protein eIF-4E of Saccharomyces cerevisiae. Molecular and cellular biology 8(8): 3556-3559. http://www.ncbi.nlm.nih.gov/pubmed/?term=3062383
47. Naranda T, MacMillan SE, Hershey JW (1994). Purified yeast translational initiation factor eIF-3 is an RNA-binding protein complex that contains the PRT1 protein. The Journal of biological chemistry 269(51): 32286-32292. http://www.ncbi.nlm.nih.gov/pubmed/?term=7798228
48. Keierleber C, Wittekind M, Qin SL, McLaughlin CS (1986). Isolation and characterization of PRT1, a gene required for the initiation of protein biosynthesis in Saccharomyces cerevisiae. Molecular and cellular biology 6(12): 4419-4424. http://www.ncbi.nlm.nih.gov/pubmed/?term=3025657
50. Bedard DP, Johnston GC, Singer RA (1981). New mutations in the yeast Saccharomyces cerevisiae affecting completion of “start”. Current genetics 4(3): 205-214. http://dx.doi.org/10.1007/BF00420500
51. Unger MW, Hartwell LH (1976). Control of cell division in Saccharomyces cerevisiae by methionyl-tRNA. Proceedings of the National Academy of Sciences of the United States of America 73(5): 1664-1668. http://dx.doi.org/10.1073/pnas.73.5.1664
52. Yu L, Pena Castillo L, Mnaimneh S, Hughes TR, Brown GW (2006). A survey of essential gene function in the yeast cell division cycle. Molecular biology of the cell 17(11): 4736-4747. http://dx.doi.org/10.1091/mbc.E06-04-0368
108. Bernstein KA, Bleichert F, Bean JM, Cross FR, Baserga SJ (2007). Ribosome biogenesis is sensed at the Start cell cycle checkpoint. Molecular biology of the cell 18(3): 953-964. http://dx.doi.org/10.1091/mbc.e06-06-0512
136. Sachs AB, Davis RW (1989). The poly (A) binding protein is required for poly (A) shortening and 60S ribosomal subunit-dependent translation initiation. Cell 58(5): 857-867. http://dx.doi.org/10.1016/0092-8674(89)90938-0
137. Prendergast JA, Murray LE, Rowley A, Carruthers DR, Singer RA, Johnston GC (1990). Size selection identifies new genes that regulate Saccharomyces cerevisiae cell proliferation. Genetics 124(1): 81-90. http://www.ncbi.nlm.nih.gov/pubmed/?term=2407608
138. Bernstein KA, Baserga SJ (2004). The small subunit processome is required for cell cycle progression at G1. Molecular biology of the cell 15(11): 5038-5046. http://dx.doi.org/10.1091/mbc.e04-06-0515