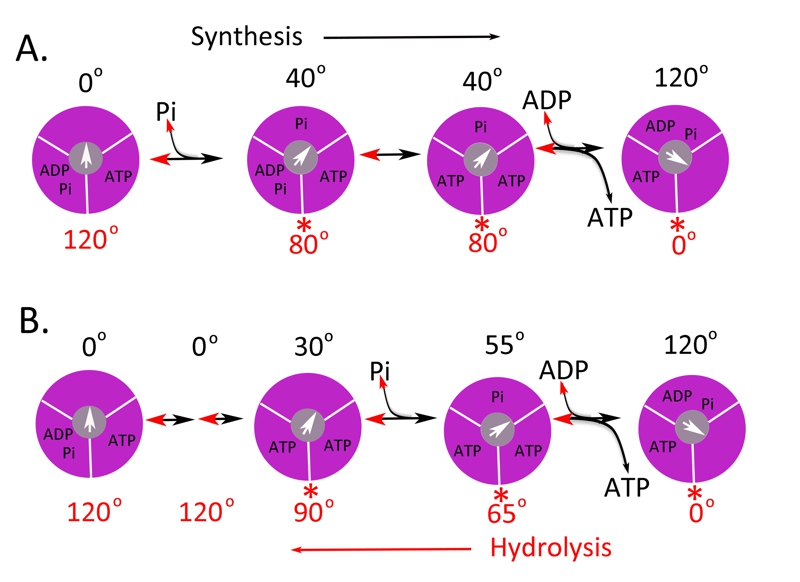
FIGURE 2: Possible scheme for site occupancy during ATP synthesis.
Shown is the one possible mechanism for ATP synthesis based on the data for the bacterial [34] and yeast [35] enzyme (A) and the human (B) F1 ATPase [41]. The direction from left to right is for ATP synthesis and right to left for ATP hydrolysis. The three binding sites are shown in magenta and the γ-subunit in grey with the arrow indicating the relative position. The asterisks (*) indicate the dwell angle observed during ATP hydrolysis.
34. Adachi K, Oiwa K, Nishizaka T, Furuike S, Noji H, Itoh H, Yoshida M, and Kinosita K, Jr. (2007) Coupling of rotation and catalysis in F1-ATPase revealed by single-molecule imaging and manipulation. Cell 130, 309-321. http://dx.doi.org/10.1016/j.cell.2007.05.020
35. Steel BC, Nord AL, Wang Y, Pagadala V, Mueller DM, and Berry RM. (2015) Comparison between single-molecule and X-ray crystallography data on yeast F1-ATPase. Scientific Reports 5, 8773. http://dx.doi.org/10.1038/srep08773
41. Suzuki T, Tanaka K, Wakabayashi C, Saita E, and Yoshida M. (2014) Chemomechanical coupling of human mitochondrial F1-ATPase motor. Nat Chem Biol 10, 930-936. http://dx.doi.org/10.1038/nchembio.1635