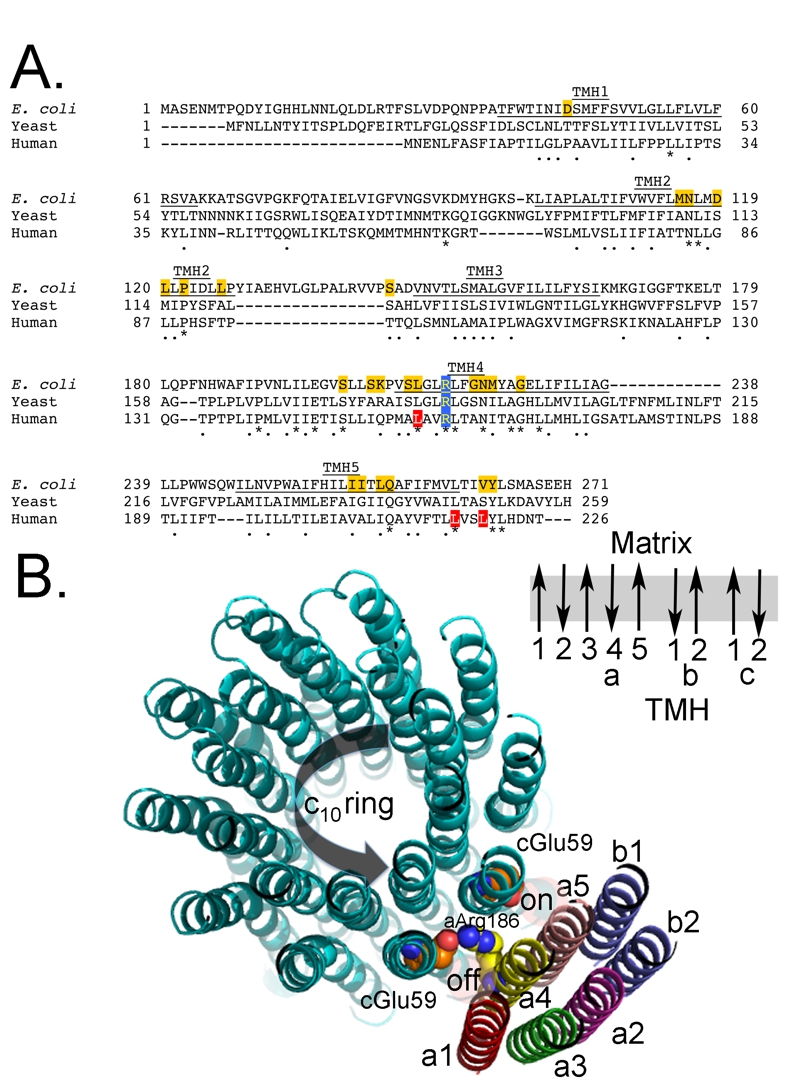
FIGURE 4: Primary sequence alignment of subunit-a.
(A) The partial primary sequence alignment of subunit-a from E. coli, yeast, and human is shown. The residues predicted to form transmembrane helices 3-5 (TMH3-5) are underlined and labeled [54, 56, 63, 145]. The highly conserved and essential R210 (E. coli numbering) is shaded blue. The residues shaded red are mutated in the discussed human diseases. The residues shaded gold are identified as important for proton movement [54, 204].
(B) Hypothetical model of the arrangement of the helices from subunits a, b, and c. The helices from subunits a (5 helices), b (2 helices) and c (20 helices) are shown along with the side chains for a-Arg1876 (yellow/blue) and c-Glu59 (red). There is very little cross-linking data positioning Helix 1 of subunit-a and thus the position is inferred from the absence, rather than presence, of cross-linking. Subunit b is thought to have a stoichiometry of 2 in the E. coli enzyme and 1 in the mitochondrial enzyme and there is 1 transmembrane helix in the bacterial subunit and 2 in the mitochondrial subunit. This view is from the matrix side of the mitochondrion. Also shown is the relative direction for deportation/protonation of c-Glu59 during ATP synthesis. The black arrow indicates the direction of rotation during ATP synthesis. Also shown is a schematic of the orientation of the transmembrane helices (TMH) for subunits a, b, and c in the membrane. Note that the helices for the subunits-a and -b are representations of a possible placement and secondary structure, while the structure of the c-ring is a crystal structure obtained from yeast [8].
8. Symersky J, Pagadala V, Osowski D, Krah A, Meier T, Faraldo-Gomez JD, and Mueller DM. (2012) Structure of the c10 ring of the yeast mitochondrial ATP synthase in the open conformation. Nat Struct Mol Biol 19, 485-491. http://dx.doi.org/10.1038/nsmb.2284
54. Dong H, and Fillingame RH. (2010) Chemical reactivities of cysteine substitutions in subunit a of ATP synthase define residues gating H+ transport from each side of the membrane. J Biol Chem 285, 39811-39818. http://dx.doi.org/10.1074/jbc.M110.175844
56. Long JC, Wang S, and Vik SB. (1998) Membrane topology of subunit a of the F1F0 ATP synthase as determined by labeling of unique cysteine residues. J Biol Chem 273, 16235-16240. http://dx.doi.org/10.1074/jbc.273.26.16235
63. Vik SB, Long JC, Wada T, and Zhang D. (2000) A model for the structure of subunit a of the Escherichia coli ATP synthase and its role in proton translocation. Biochim Biophys Acta 1458, 457-466. http://dx.doi.org/10.1016/s0005-2728(00)00094-3
145. Vik SB, and Ishmukhametov RR. (2005) Structure and function of subunit a of the ATP synthase of Escherichia coli. J Bioenerg Biomembr 37, 445-449. http://dx.doi.org/10.1007/s10863-005-9488-6
204. Steed PR, and Fillingame RH. (2009) Aqueous accessibility to the transmembrane regions of subunit c of the Escherichia coli F1F0 ATP synthase. J Biol Chem 284, 23243-23250. http://dx.doi.org/10.1074/jbc.m109.002501