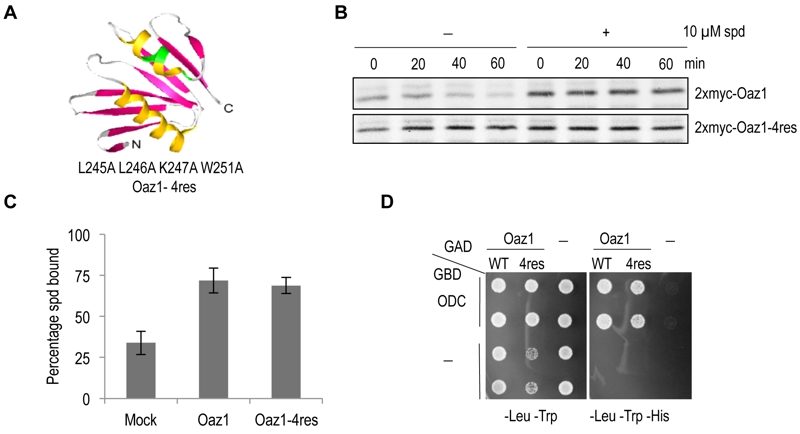
FIGURE 1: The antizyme mutant Oaz1-4res is metabolically stable in yeast cells.
(A) Ribbon diagram of the NMR structure of rat antizyme [PDBcode 1ZO0] with β-sheets shown in pink and α-helices in yellow. Four amino acid changes (L245A L246A K247A W251A) were introduced into the yeast antizyme homologue at corresponding positions (indicated in green). The diagram was prepared using 3D molecular viewer.
(B) Pulse-chase analysis of 35S-radiolabelled Oaz1 (upper panel) or Oaz1-4res (lower panel) in cells grown in the presence or absence of 10 µM spermidine (spd) showing that the stability of Oaz1-4res is independent of polyamine addition.
(C) Spermidine binding assay showing the retention of [3H]-spermidine by 6His-Oaz1 or 6His-Oaz1-4res. Error bars, s.d.; n = 3.
(D) Yeast two-hybrid analysis showing the interaction of Gal4 DNA binding domain (GBD) fused to ODC (GBD-ODC) with Gal4 transcription activation domain (GAD) fused to either Oaz1 or Oaz1-4res (GAD-Oaz1 or GAD-Oaz1-4res). Interaction of the two separated Gal4 domains via the polypeptides fused to them leads to reconstitution of the Gal4 transcriptional activator which controls expression of the HIS3 gene in the reporter strain used. Interaction-mediated functional reconstitution of Gal4 can be monitored as growth on medium lacking histidine in this strain.