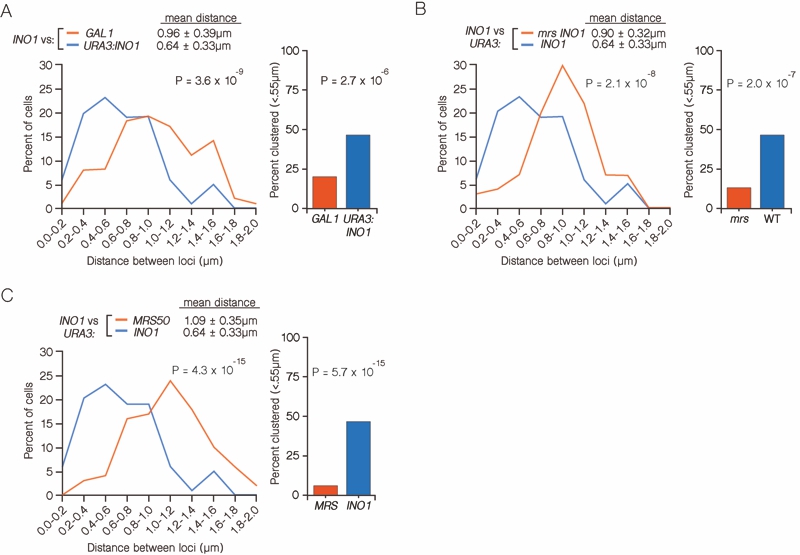
FIGURE 3: INO1 interchromosomal clustering during memory is specific and MRS-dependent. (A-C) Haploid cells having the endogenous INO1 gene marked with the TetO and either URA3 (A-C) or GAL1 (A) marked with the LacO, expressing GFP-TetR and mRFP-LacI [8] were grown under INO1 memory (activating → repressing, 3h) conditions, fixed and processed for immunofluorescence against GFP and mRFP. Left: The distribution of distances between the two loci in ~ 100 cells, binned into 0.2 µm bins. P values were calculated using a Wilcoxon Rank Sum Test. Right: the fraction of cells in which the two loci were ≤ 0.55 µm. P values were calculated using a Fisher Exact Test. Note that the data used to generate the distribution for the control of INO1-TetO vs. URA3:INO1-LacO is the same in all three panels. The combinations tested were INO1-TetO vs. URA3:INO1-LacO (A-C), or INO1-TetO vs. GAL1-LacO (A), INO1-TetO vs. URA3:mrsINO1-LacO (B) and INO1-TetO vs. URA3:MRS50-LacO (C).
8. Brickner DG, Cajigas I, Fondufe-Mittendorf Y, Ahmed S, Lee PC, Widom J and Brickner JH (2007). H2A.Z-mediated localization of genes at the nuclear periphery confers epigenetic memory of previous transcriptional state. PLoS Biol 5:e81. http://dx.doi.org/10.1371/journal.pbio.0050081